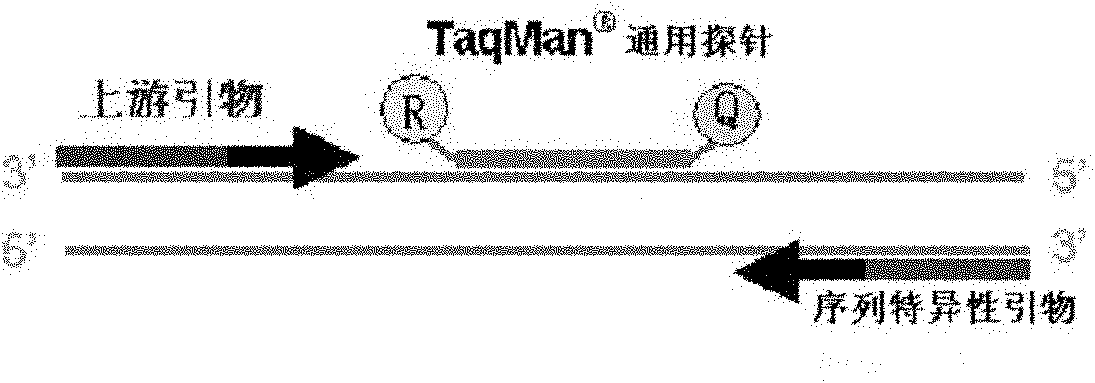
Method and kit for detecting drug resistance of mycobacterium tuberculosis
A Mycobacterium tuberculosis, drug resistance technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of high cost of LiPA, wrong nucleotides, inability to obtain drug resistance information, etc., to improve sensitivity and Effect of Amplification Efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1, Primers and Probes for Detecting Mycobacterium tuberculosis drug-resistant gene mutations
[0063] 1. Design and synthesis of primers
[0064] In order to specifically and efficiently amplify the target gene in the nucleic acid sample to be tested, the whole genome DNA (BX842578) sequence of Mycobacterium tuberculosis (MTB) was retrieved from GenBank, and the 759807-763325th nucleoside of rpoB (NC_000962 acid), katG (NC_000962 2153889-2156111 nucleotides), inhA (NC_000962 1674202-1675011 nucleotides) gene polymorphism sites, according to the general principle of primer design, using Primer Premier5.0 and Oligo6 .0 software designed and synthesized the forward and reverse primers used in real-time fluorescent PCR.
[0065] Primer set 1:
[0066] rpoB_comF2: 5'AGCCGATGACCACCCAGGAC 3' (sequence 1)
[0067] 533_R3: 5'AGACCGCCGAGCCCCG 3' (sequence 2)
[0068] 531_Ri2: 5'CCGAGCCCCAGCGCCA 3' (sequence 3)
[0069] 526_R3: 5'CGGCAGTCGGCGCTTGTC 3' (sequence 4)
...
Embodiment 2
[0083] Embodiment 2, the condition exploration of real-time fluorescent PCR
[0084] 1. Template preparation
[0085] According to the 759807-763325 nucleotides of NC_000962, the 1674202-1675011 nucleotides of NC_000962, and the 2153889-2156111 nucleotides of NC_000962, the rpoB gene, inhA gene and katG gene were artificially synthesized respectively, and the artificially synthesized rpoB gene was sequenced The nucleotide sequences of the , inhA gene and katG gene are respectively consistent with the 759807-763325 nucleotides of NC_000962, the 1674202-1675011 nucleotides of NC_000962, and the 2153889-2156111 nucleotides of NC_000962.
[0086] The artificially synthesized rpoB gene, inhA gene, and katG gene were respectively connected to pGEM-T Easy Vector (Promega Company, catalog number: A1380) to obtain wild-type plasmid rpoB, wild-type plasmid inhA, and wild-type plasmid katG, respectively.
[0087] Sequenced:
[0088] The wild-type plasmid rpoB is a plasmid obtained by lin...
Embodiment 3
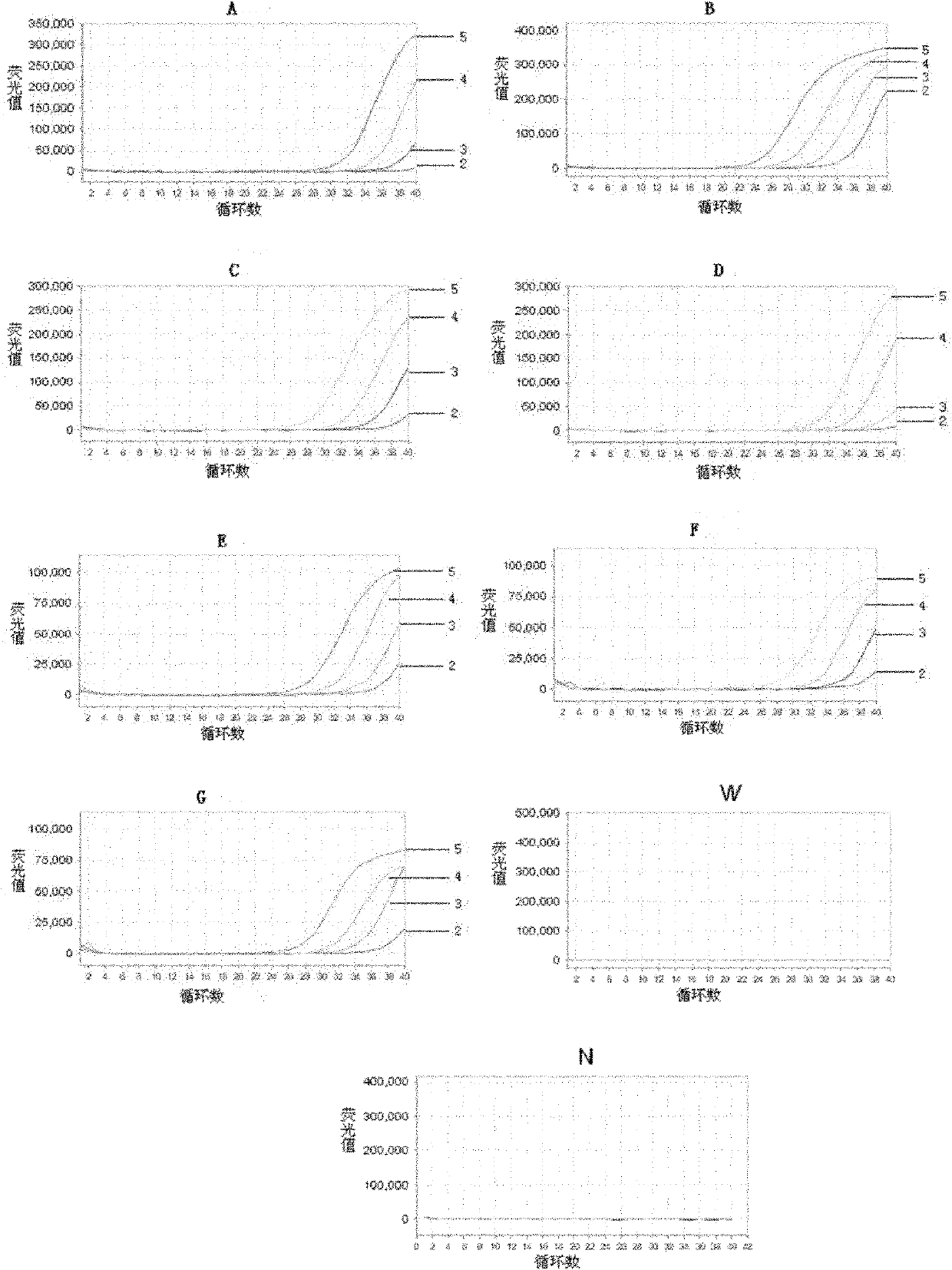
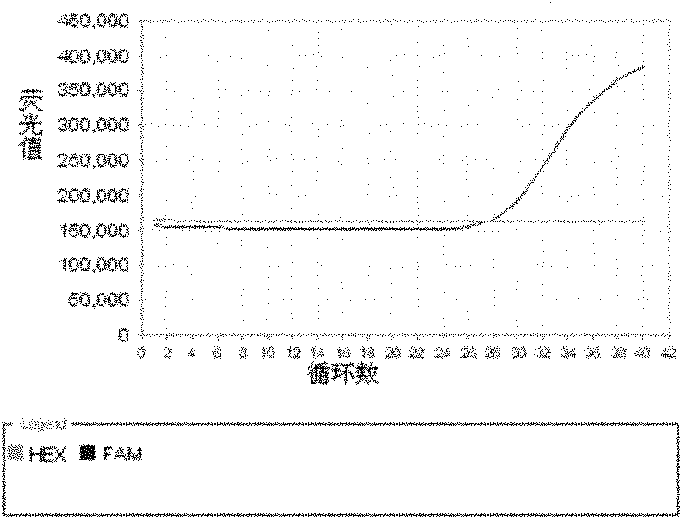
[0117] Embodiment 3, detect the drug resistance of Mycobacterium tuberculosis in the sample
[0118] 1. Real-time fluorescent quantitative PCR detection of different biological samples
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com