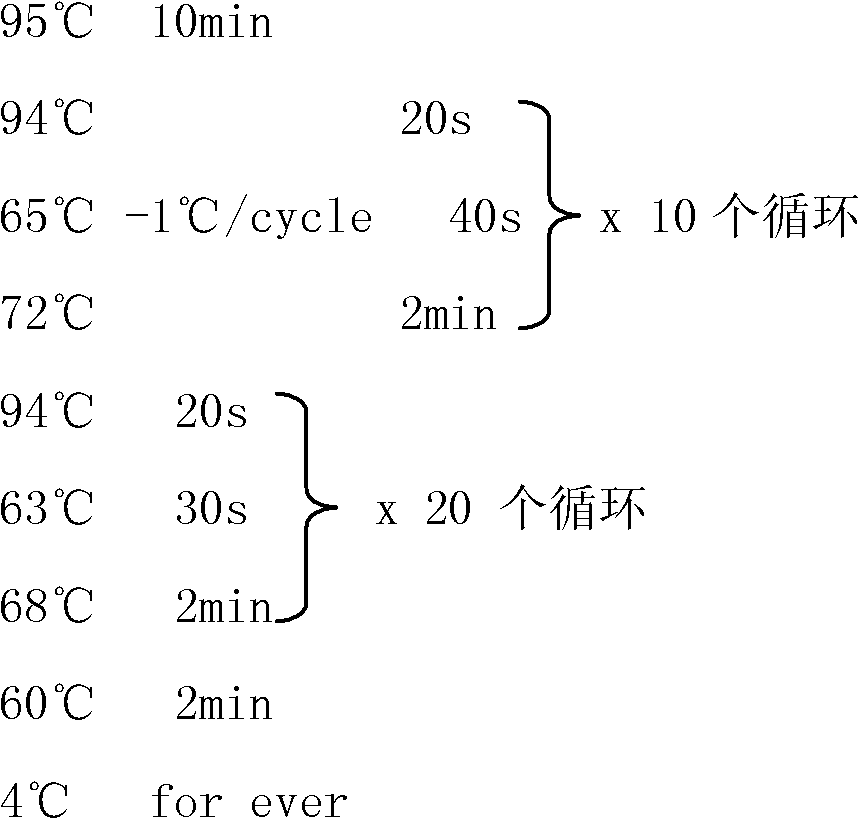Application of Y-STR (Y chromosome-short tandem repeat)
A Y-STR, individual technology, applied in the biological field, can solve the problems of difficult genetic relationship and low mutation rate of Y-STR, and achieve the effect of improving the resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
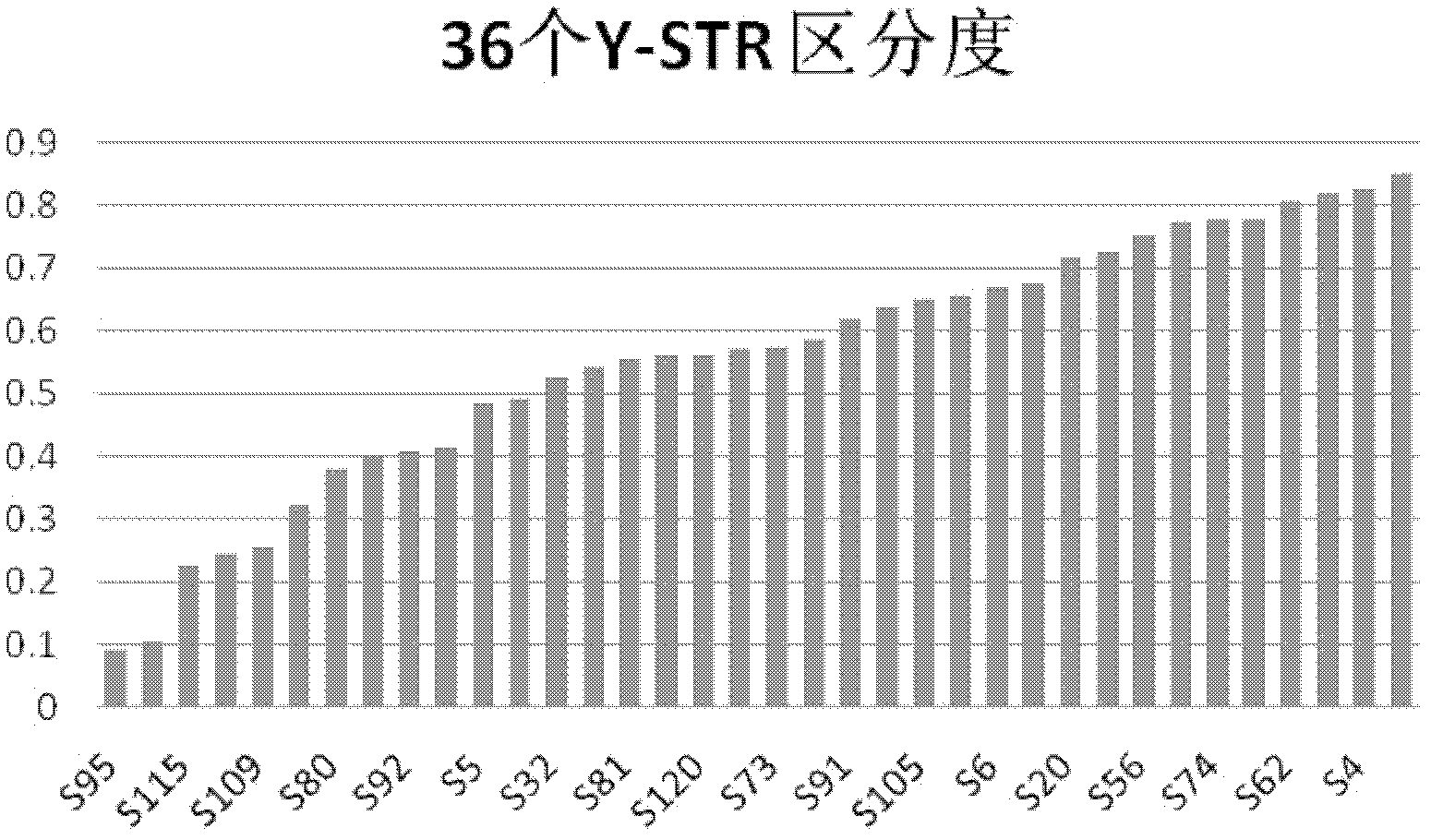
[0029] Example 1 screened out 36 new Y-STR sites
[0030] 1. Prediction of Y-STR
[0031] (1) Checked and sorted out STR information in biological public databases, and obtained 201 STR information reported by NCBI (as of 2011-11);
[0032] (2) Bioinformatics predicts STR, and obtains 128 Y-STR sites with repeat units repeated more than 10 times;
[0033] (3) Compare the number of repetitions of these STR sites in the three genomes (refercese, HuRef, Celera) in the NCBI database, and obtain information on STR sites with large differences in the number of repetitions;
[0034] (4) Combined with the literature, biological databases and bioinformatics predictions, according to the sequence characteristics of the actual Y-STRs, the reported sites with DYS numbers were removed, and 103 Y-STRs predicted by bioinformatics were screened out, named as They are respectively named S1, S2...S128, S128 according to the size of their positions on the Y chromosome. The 36 new Y-STR loci s...
Embodiment 2
[0059] Example 2 Clarify the genetic relationship by detecting 36 Y-STRs
[0060] Thirty-nine male samples were collected from different regions, some of whom were close relations such as father and son, uncle and nephew. By detecting the 36 Y-STRs screened out in Example 1 above, their genetic relationship was clarified.
[0061] The results of the data show that through these 36 newly discovered Y-STR loci, the relative relationship between father and son or uncle and nephew in these 39 male samples from different regions is clearly clarified (see Table 2 on the next page). The common Y-STR loci -STR is difficult to distinguish between uncles and nephews or individuals who are relatively close relatives. In this aspect, the detection accuracy of the 36 Y-STR sites newly discovered by the present invention is several orders of magnitude higher than that of the commonly used Y-STR sites.
[0062] Table 2 Y-STR genetic polymorphism data analysis of 39 male samples
[0063] N...
Embodiment 3
[0065] Example 3 Individual identification by detecting 36 Y-STRs
[0066] Collect random 150 male samples (the samples come from the male samples of random physical examination in Shanghai Ren'ai Hospital), and use the 36 Y-STR new sites of the present invention to detect whether it can achieve the purpose of distinguishing each person.
[0067] The experimental results show that in 150 male samples, 150 haplotypes were found (see the test data of 150 random samples in Table 3 below for detailed haplotype information), which can 100% distinguish the collected random samples, further proving that The 36 Y-STRs newly discovered by the present invention have a very high degree of discrimination.
[0068] Table 3 Test data of 150 random samples
[0069]
[0070]
[0071] Note: In Table 3, because the samples come from random physical examination samples, the sample numbers follow the hospital numbers.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com