Genome scale metabolic network model-based metabolic engineering design prediction method
A genome-scale, metabolic network technology, applied in the field of metabolic engineering design prediction based on the genome-scale metabolic network model, can solve problems such as inability to simulate gene sequences, no metabolic engineering design, and inability to determine the optimal synthetic pathway
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] The metabolic engineering design prediction method based on the genome-scale metabolic network model of the present invention will be described in detail below with reference to the embodiments and the accompanying drawings.
[0019] The realization of the metabolic engineering design prediction method based on the genome-scale metabolic network model of the present invention firstly ensures that the whole-genome scale metabolic network model as the basis has high quality. And, the present invention utilizes COBRA toolkit to carry out programming calculation.
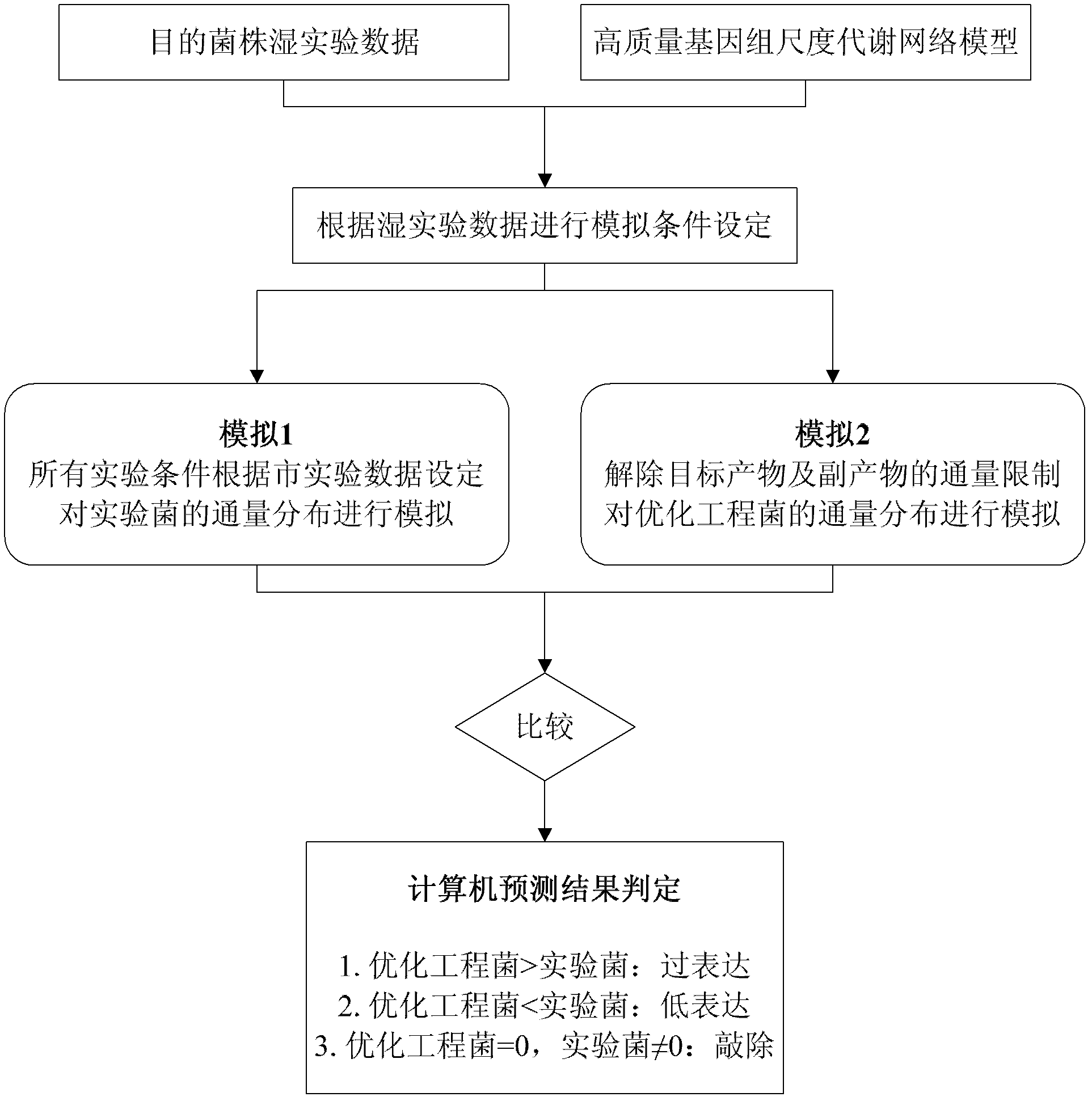
[0020] Such as figure 1 As shown, the metabolic engineering design prediction method based on the genome-scale metabolic network model of the present invention includes the following steps:
[0021] 1) Obtain the experimental data of the target bacteria and the high-quality genome-scale metabolic network model respectively;
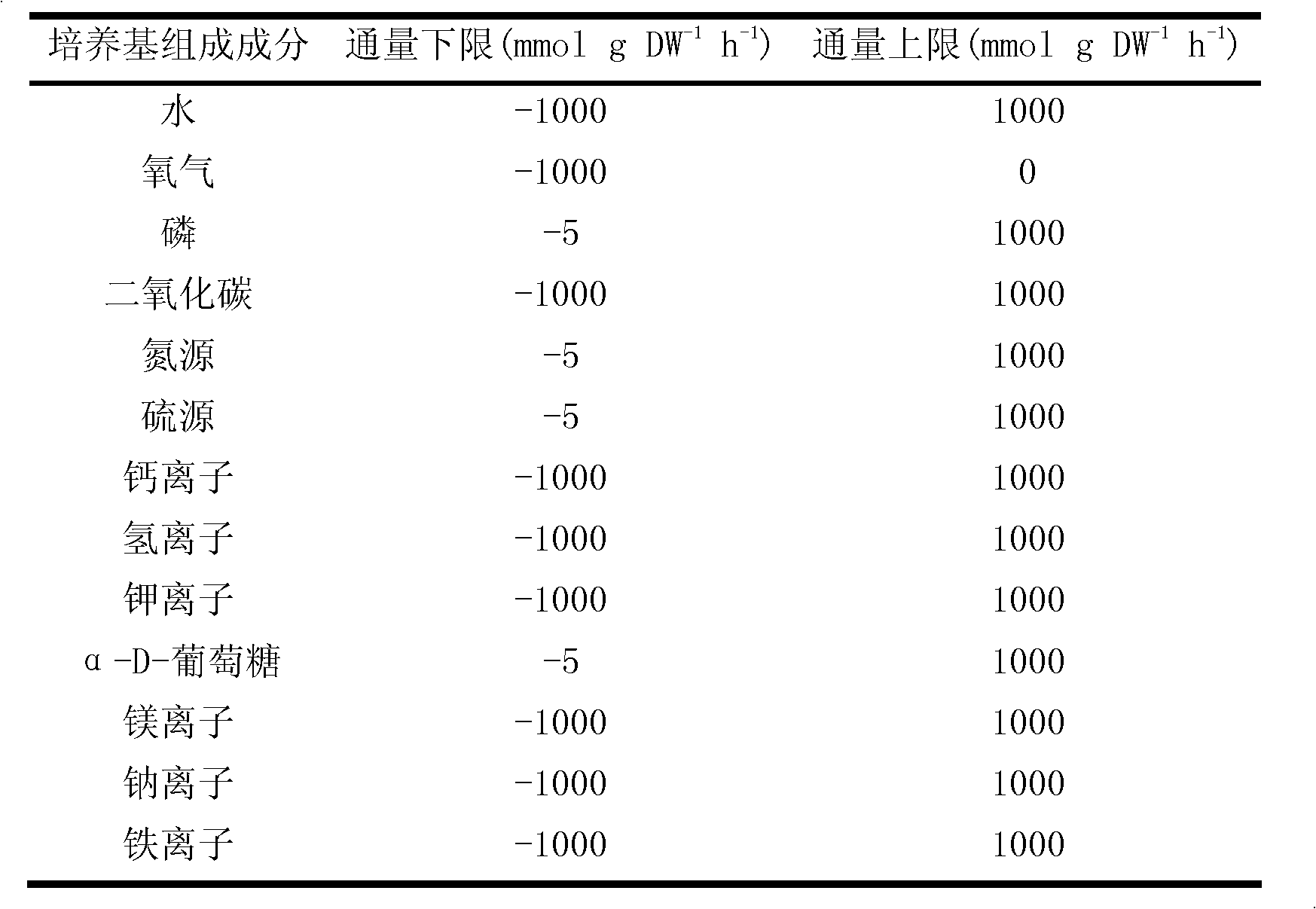
[0022] 2) Set the simulation conditions according to the wet test data,
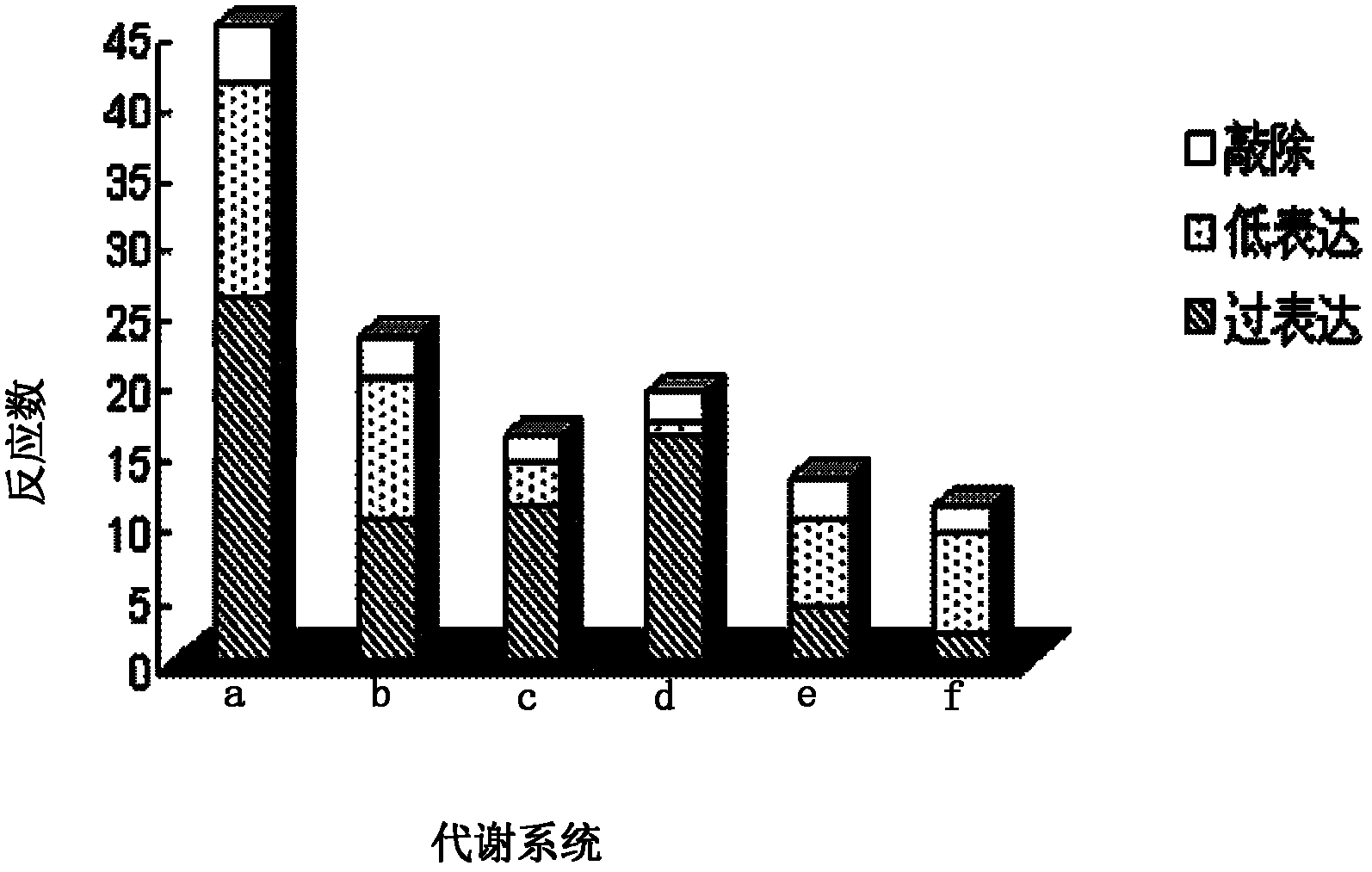
[0023] The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com