Materials and methods for isothermal nucleic acid amplification
A technology for isothermal amplification and target nucleic acid, which is applied in the field of thermal amplification of target RNA, and can solve problems such as lack of proofreading ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1: Use of PYROPHAGE 3173 as a replacement for RT enzyme in RT-HAD
[0060] PYROPHAGE 3173 DNA polymerase has several advantages over Transcriptor and Thermoscript reverse transcriptases. For example, Thermoscript and Transcriptor are known to have limited activity in HAD buffer at 65°C. PYROPHAGE 3173 DNA polymerase was tested to determine if it could replace these reverse transcriptases in a one-step isothermal RT-HAD amplification.
[0061] Briefly, a 25 μL reaction mixture was created containing: (1) 0, 10, or 100 copies of an in vitro transcribed, synthetic RNA comprising the Chlamydia trachomatis cryptic plasmid RNA (ct-RNA) ( GenBank accession number X06707) (SEQ ID NO:1); (2) forward primer 5'-ATC GCA TGC AAG ATA TCG AGT ATG CGT-3'(SEQ ID NO:2) and reverse primer 5'-CTC ATA ATT AGC AAG CTG CCT CAG AAT-3' ("ct-orf primer") (SEQ ID NO:3); (3) 2.5 U of Thermoscript, Thermo-X, Transcriptor, or Pyrophage 3173; (4) 2 U of Bst polymerase; and (5) 1 U of uvrD ...
Embodiment 2
[0065] Example 2: Use of PYROPHAGE 3173 as a replacement for both RT and DNA-dependent DNA polymerases
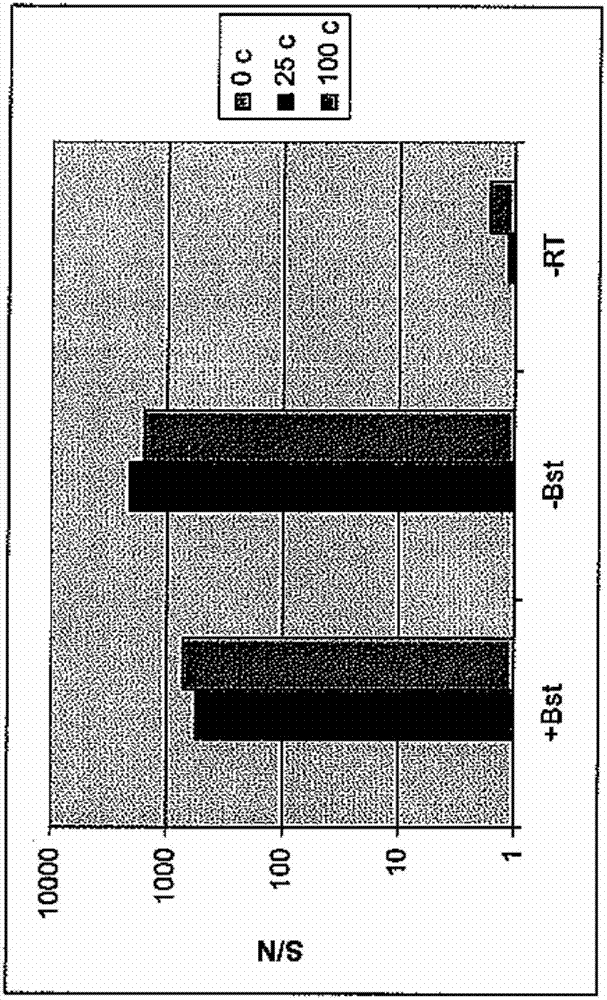
[0066] PYROPHAGE 3173 DNA polymerase was tested to determine if it could replace both reverse transcriptase and DNA-dependent DNA polymerase in a one-step isothermal RT-HAD amplification. Briefly, a 25 μL reaction mixture was created containing: (1) 0, 25, or 100 copies of ct-RNA; (2) ct-orf primers; (3) 2.5 U of Pyrophage 3173; (4) 0 U or 2 U Bst polymerase; and (5) 1 UuvrD helicase. The reaction mixture contained the reagents at the final concentrations listed in Table 1. Amplification was performed at 62°C or 65°C for 75 minutes. exist figure 2 show result. As can be seen, PYROPHAGE 3173 is able to replace both reverse transcriptase and DNA-dependent DNA polymerase in a one-step isothermal RT-HAD.
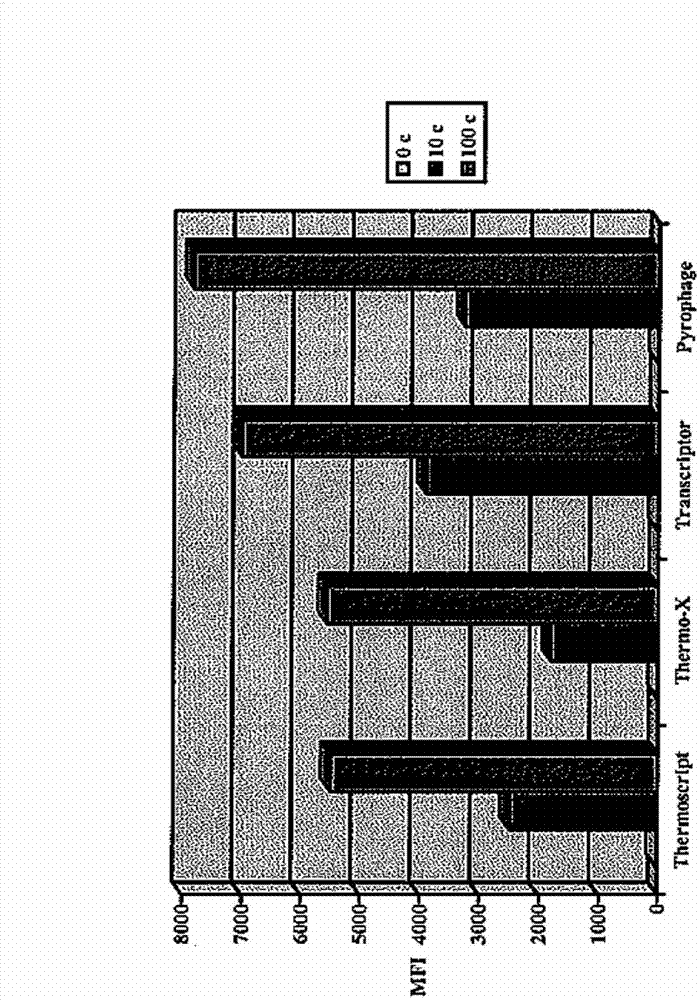
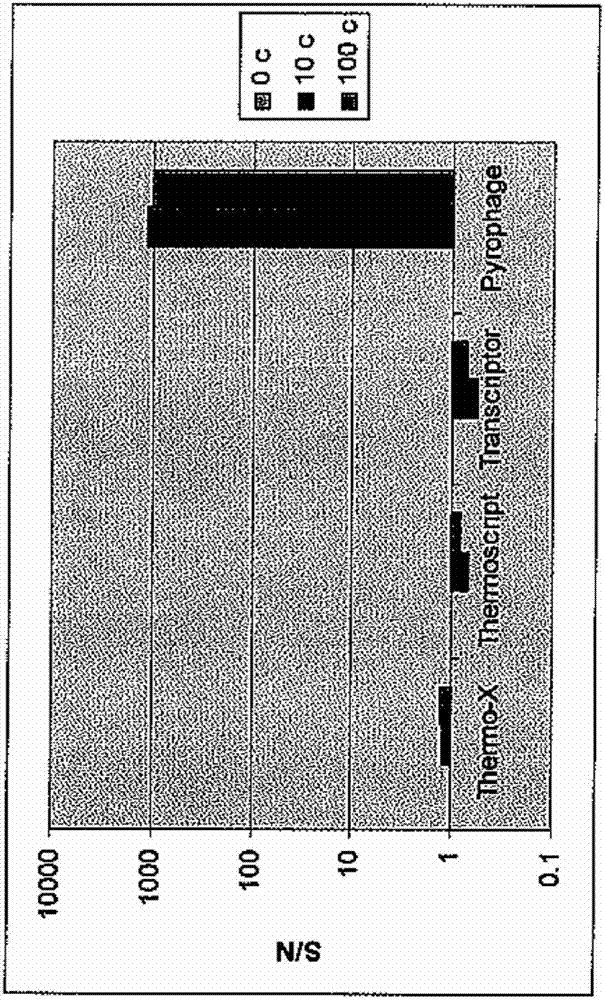
[0067] PYROPHAGE 3173 DNA polymerase was also compared to other enzymes with reverse transcriptase activity in its ability to perform RT-HAD in the absence of different...
Embodiment 3
[0068] Example 3: DNA Amplification Using PYROPHAGE 3173
[0069] target amplification . Use of HPV16 DNA as target DNA in the HAD assay. Double-stranded DNA targets were denatured in 5 μl of 0.1 M NaOH at 65°C for 10 min. Then, an equal volume of 0.2M Hepes was added to neutralize the denatured target. 15 μl premix and 25 μl amplification mix were added to the targets and incubated at 65°C for 1.5 hours. Table 2 lists the master mix and amplification mix components.
[0070] Amplicon Detection . The HAD product (5 μL) was transferred to a U-bottom hybridization plate and then diluted in 5 μl of 1X Denaturing Reagent (Digene HC2DNR, Qiagen Gaithersburg, Inc., Gaithersburg, MD). The plate was then sealed and shaken at 1100 rpm for 30 seconds in a Digene shaker and incubated at room temperature for 15 minutes. Hybridization diluent (5 μl 1X hc2 Probe Diluent, Qiagen Gaithersburg, Inc., Gaithersburg, MD) was added and the plate was resealed and shaken at 1100 rpm for 30 s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com