Compositions and methods for reverse transcription of nucleic acid molecules
a nucleic acid molecule and reverse transcription technology, applied in the field of molecular and cellular biology, can solve the problems of inefficient use of unmodified rt to catalyze reverse transcription, heterodimeric rt formation, etc., and achieve the effect of reducing or substantially reducing the activity of rnase h
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
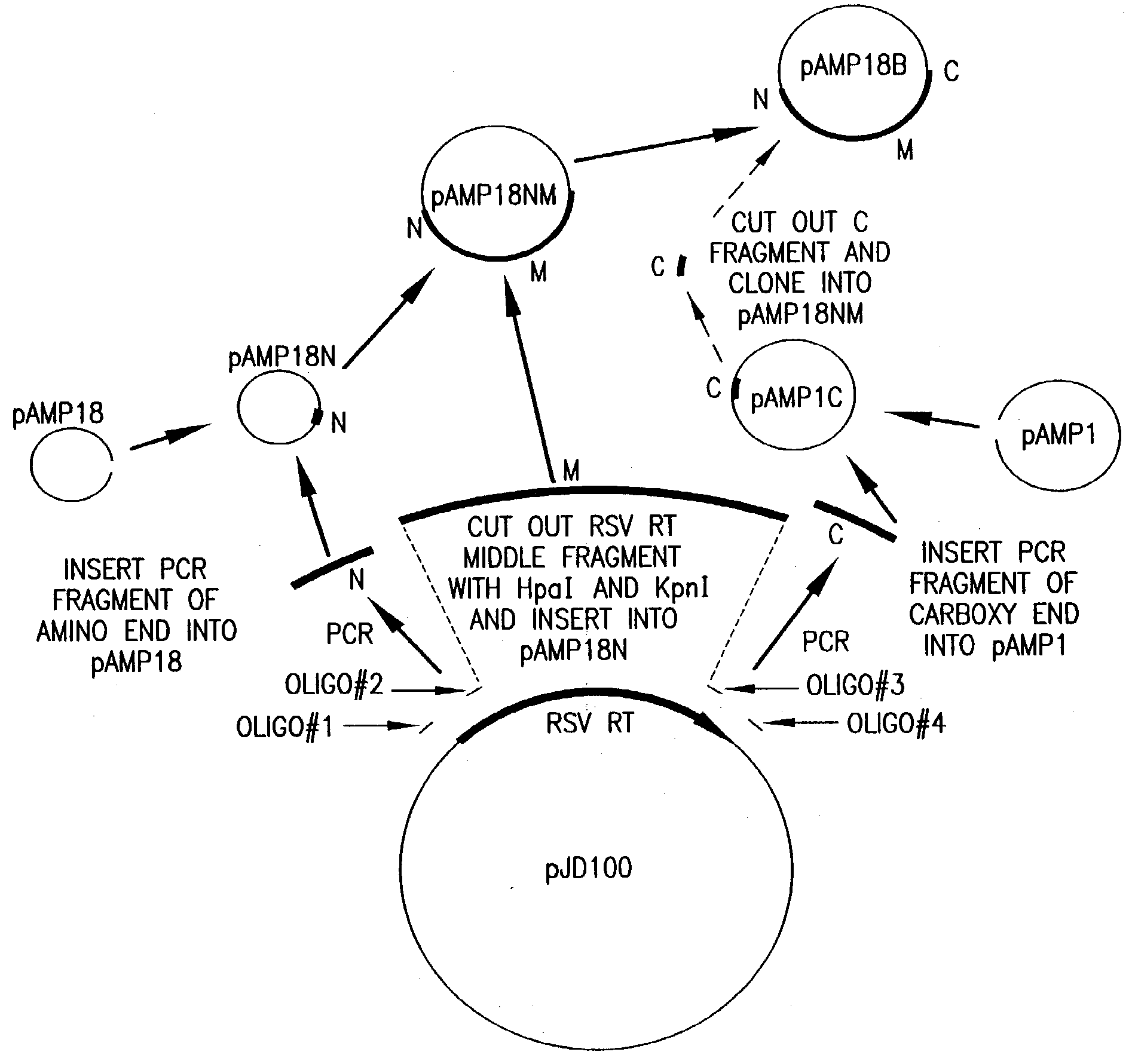
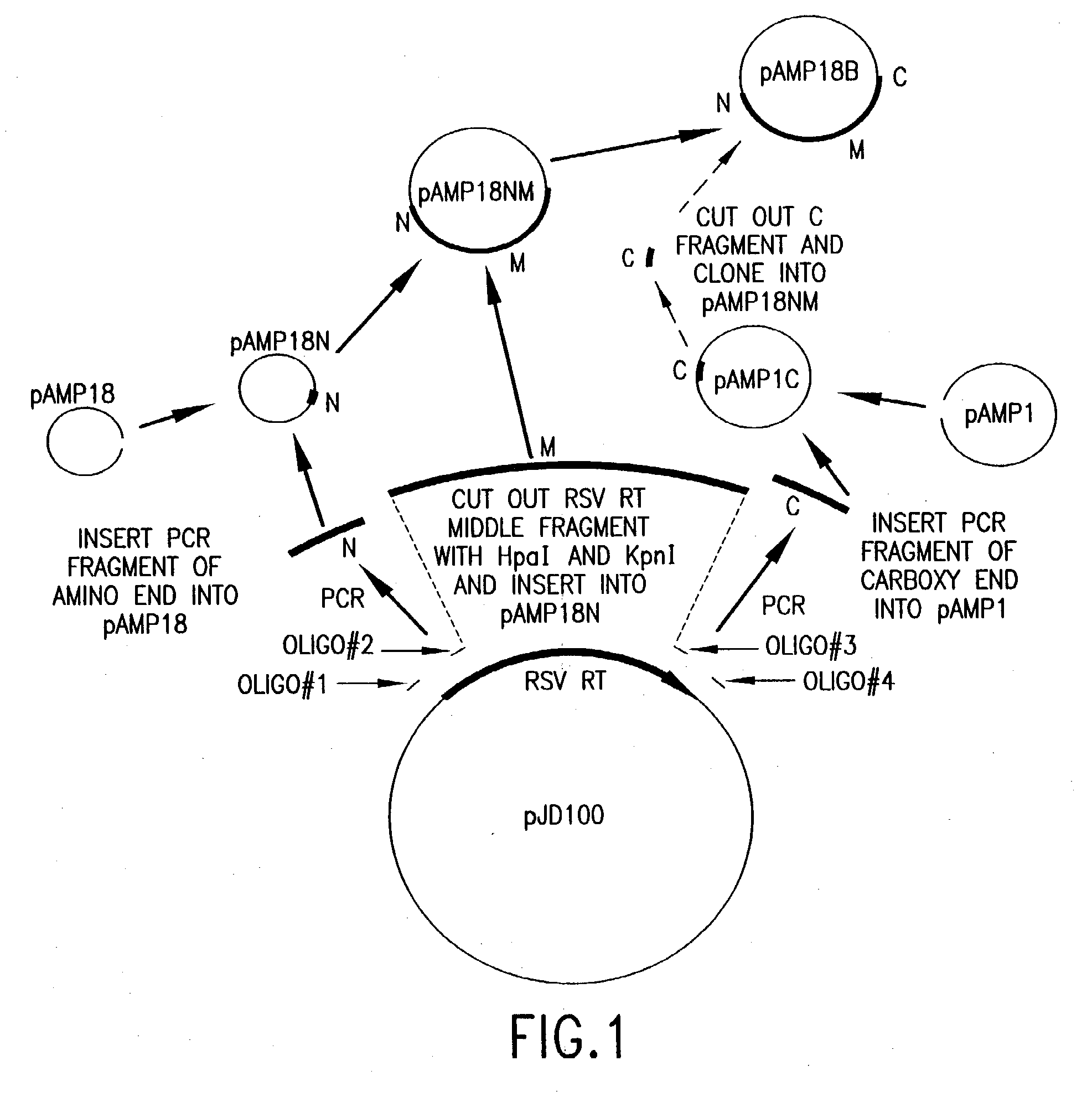
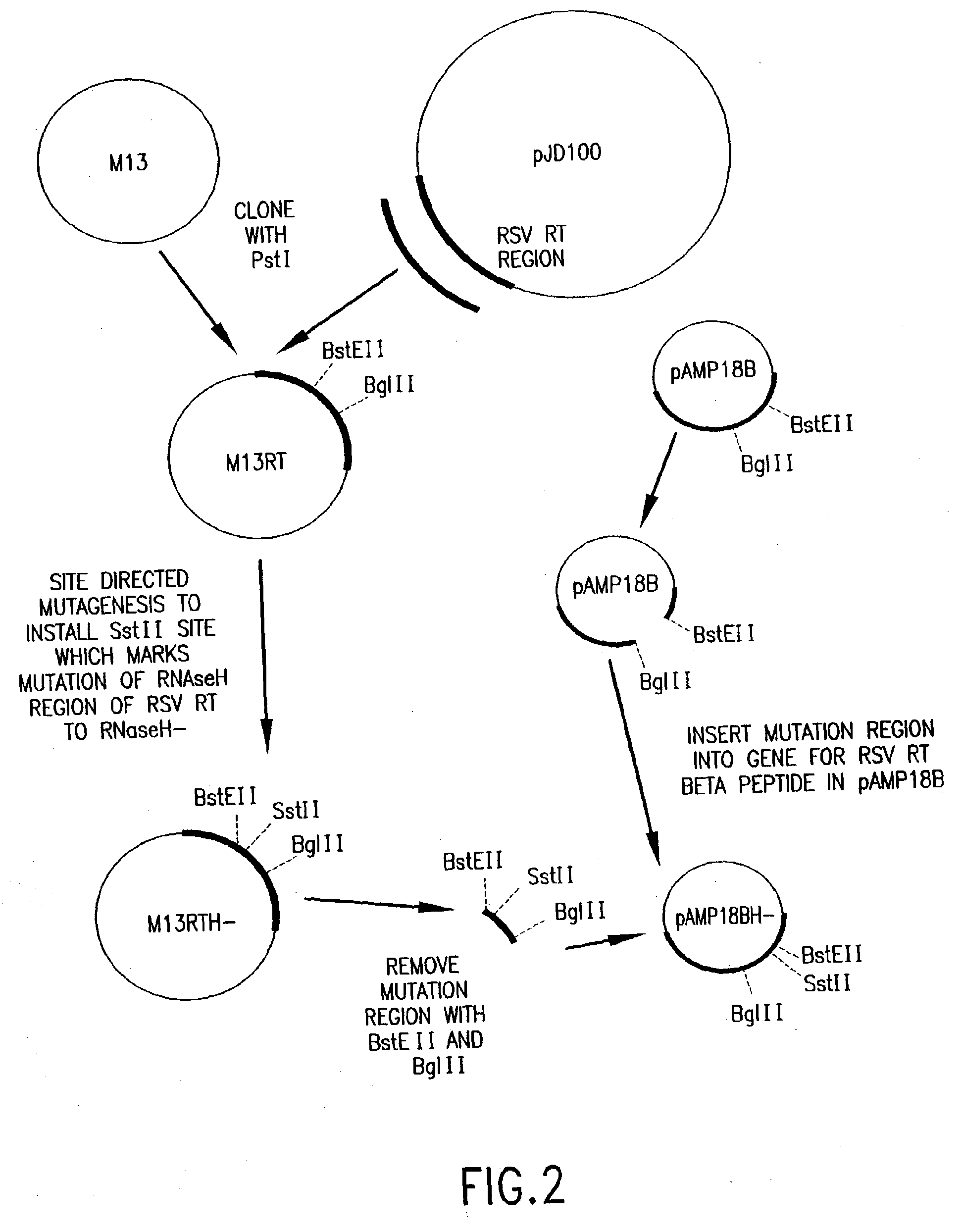
Cloning and Expression of RSV RNase H− RT
[0153] General Methods
[0154] RSV H− RT is a cloned form of retrovirus RT, in which both the α and the β subunits are mutated by a single amino acid change to eliminate RNase H activity. An RSV RT exhibiting substantially reduced RNase H activity is also produced when only the α subunit is mutated to substantially reduce RNase H activity (with the β subunit not being mutated in the RNase H domain). Mutations and plasmid constructions were conducted using standard molecular biology methods (see, e.g., Sambrook, J., et al., Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor, N.Y.: Laboratory Press (1989)), modified as described below.
[0155] Plasmid preparation. Plasmid preparations from 1 ml E. coli cultures were made by the alkaline lysis procedure (Sambrook, J., et al., Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor, N.Y.: Laboratory Press (1989)). From 10 ml cultures, the preparation was additionally t...
example 2
Isolation of RSV RNase H− RT
[0182] To provide purified recombinant RSV H− RT, cloned RSV H− RT was overexpressed in cultured insect cells as described in Example 1 and purified by affinity and ion-exchange chromatography. The RSV RT produced comprises the α and β subunits. Isolation of RSV RT provides a substantially pure RSV RT in which contaminating enzymes and other proteins have been substantially removed, although such contaminants need not be completely removed.
[0183] Buffers. The pH of all buffers was determined at 23° C., and buffers were stored at 4° C. until use. Crack Buffer contained 50 mM Tris-HCl (pH 7.9), 0.5 M KCl, 0.02% (v / v) Triton X-100 and 20% (v / v) glycerol. Just before use, the following protease inhibitors (Boehringer Mannheim; Indianapolis, Ind.) were added to Crack Buffer at the final concentrations indicated: leupeptin (2 mg / ml), Pefabloc (48 μg / ml), pepstatin A (2 μg / ml), benzamidine (800 μg / ml) and PMSF (50 μg / ml). Buffer A contained 20 mM Tris-HCl (pH ...
example 3
Preparation of Full-Length cDNA Molecules
[0188] Enzymes. SuperScript II RT (SS II RT), a cloned form of Moloney murine leukemia virus (M-MLV) RT lacking demonstrable RNase H activity (i.e., an “RNase H− RT”), was from Life Technologies, Inc. (Rockville, Md.). M-MLV RT, a cloned murine RT with full RNase H activity (i.e., an “RNase H+ RT”), was also from Life Technologies, Inc. AMV RT, an RNase H+ uncloned form of avian myeloblastosis virus RT, was from Seikagaku America, Inc. RSV H− RT was prepared as described in Examples 1 and 2. Recombinant Tth DNA polymerase, a cloned, thermophilic DNA polymerase from Thermus thermophilus with reverse transcriptase activity, was from Perkin Elmer.
[0189] Synthetic mRNA. A 7.5 kilobase (Kb) synthetic mRNA with a 120-nucleotide 3′ poly(A) tail (Life Technologies, Inc.; Rockville, Md.) was used as template to test the efficiency of various enzymes alone or in combination.
[0190] cDNA Synthesis Reaction Mixtures. Reaction mixtures (20 μl each) cont...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com