Method for identifying new generations breeding prawn varieties
An identification method and a new variety of technology, applied in the field of molecular biology, can solve problems such as single source, achieve the effect of improving accuracy, reducing identification cost, and improving identification accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Identification process:
[0070] 1. Randomly select 71 samples of new species of prawns selected and bred through generations, and take 60 representative shrimps of Chia prawns, Tongwei shrimps and Fujian second-generation shrimps that are widely used in the market as control samples, and extract the samples of successive generations. Breeding samples of new species of prawns and DNA of control samples.
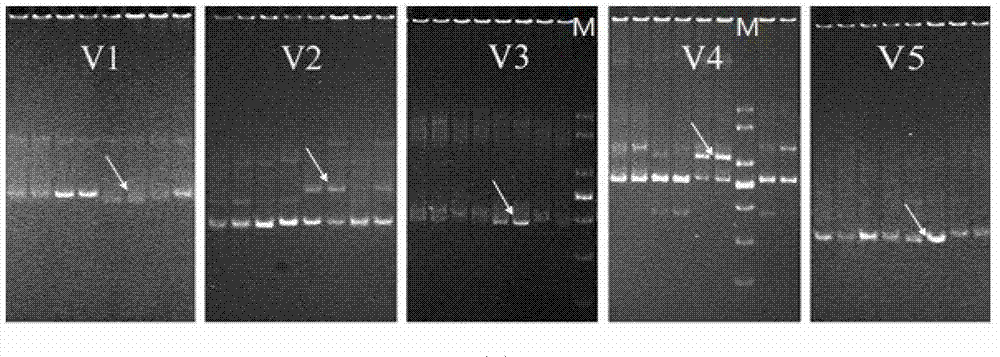
[0071] 2. Use the primer pair for amplifying the 14950-15583 segment of the mitochondrial control region of Litopenaeus vannamei to amplify the 71 successive generations of selected shrimp samples obtained above and randomly select 104 control sample DNAs from the control samples;
[0072] CRF: 5'-ACCATTGACCTAAAAGTGAAAGAAC-3'
[0073] CRR: 5'-ATCAATAAATATAAATTAACTACGC-3'
[0074] Amplify the sequence with a length of 634bp, and use CRF and CRR primers for bidirectional sequencing to splice and remove low-quality sequences to obtain the results.
[0075] 3. Then use ...
Embodiment 2
[0089]1. Randomly select 50 new varieties of prawns bred through generations and randomly purchase 51 new varieties of prawns that were not bred through generations from the market. Genomic DNA was extracted using the Tiangen Marine Animal Genome Extraction Kit, and the operation was carried out according to the instructions. The DNA concentration obtained was determined.
[0090] 2. Dilute the extracted genomic DNA to 40ng / μl and use primers to amplify the mitochondrial genome control region
[0091] F:ACCATTGACCTAAAAGTGAAAGAAC
[0092] R: ATCAATAAATATAAATTAACTACGC
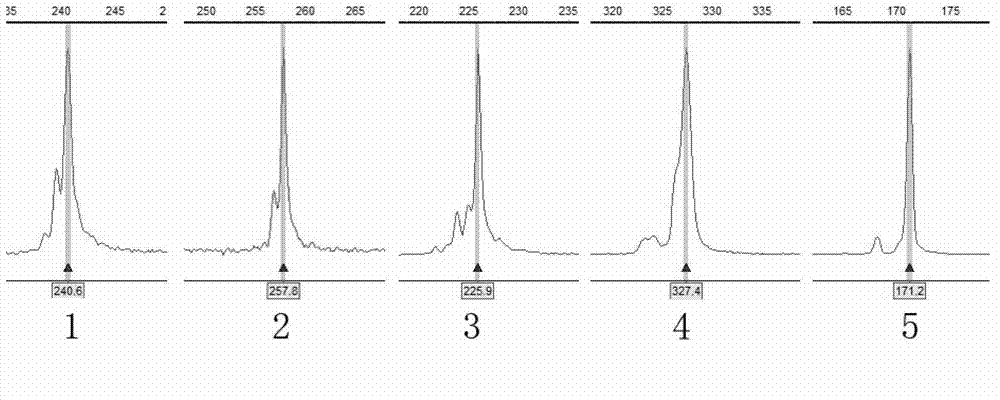
[0093] The mitochondrial genome control region sequence was amplified to obtain a 634bp sequence, and the sequence results were obtained after bidirectional sequencing using CRF and CRR primers to remove low-quality sequences, and the multiple sequence alignment results were obtained through the Clustalw program in the Bioedit software, and the results were imported into the DNAsp software Obtain haplotype info...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com