Method for detecting mycobacterium tuberculosis and nontuberculous mycobacteria using duplex real-time polymerase chain reaction and melting curve analysis
A technology of mycobacterium tuberculosis and mycobacteria, which is applied in the field of detection of mycobacterium tuberculosis and non-tuberculosis mycobacteria by using dual real-time polymerase chain reaction and melting curve analysis to achieve the effect of efficient clinical diagnosis means
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1: Isolation and Detection of Mycobacterium Tuberculosis Complex and Nontuberculous Mycobacterium 1
[0073] 1. Detection target and primer design
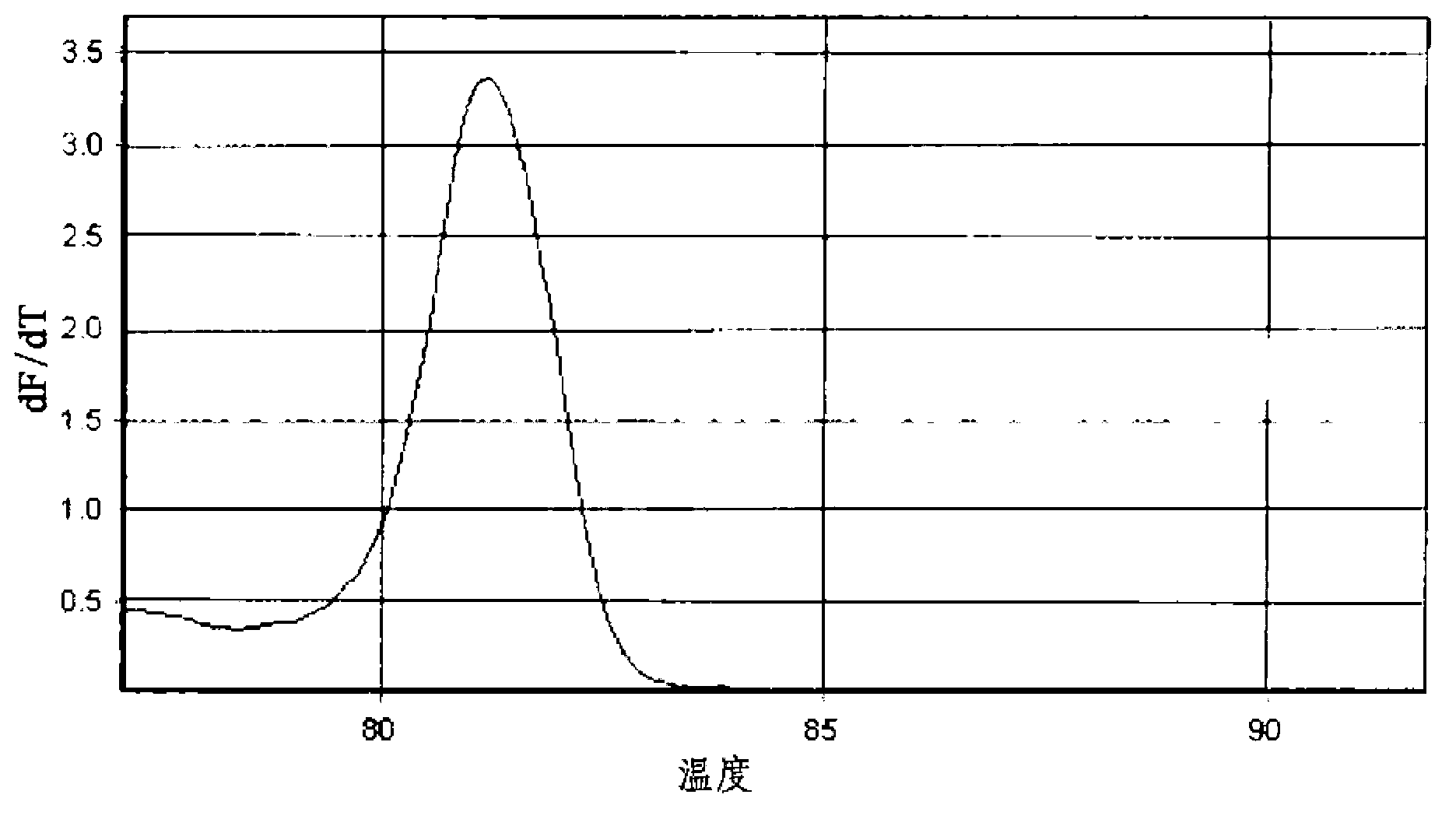
[0074] The target genes to be detected by PCR and melting curve analysis were the IS6110 gene of the Mycobacterium tuberculosis complex (Mycobacterium tuberculosis, M. 16S rRNA gene. The universal primer was used as the forward primer to amplify the 16S rRNA gene of mycobacteria. NTM-1 and NTM-2, which are characteristic of nontuberculous mycobacteria, were used as reverse primers. These primers for target gene detection were designed using the Primer3 program.
[0075] (1) Mycobacterium tuberculosis complex (MTC)
[0076] 1) Target gene: IS6110
[0077] 2) Primers
[0078] a. Forward primer: 5'-cgaactcaaggagcacatca-3' (SEQ ID NO: 1)
[0079] b. Reverse primer: 5'-agtttggtcatcagccgttc-3' (SEQ ID NO: 2)
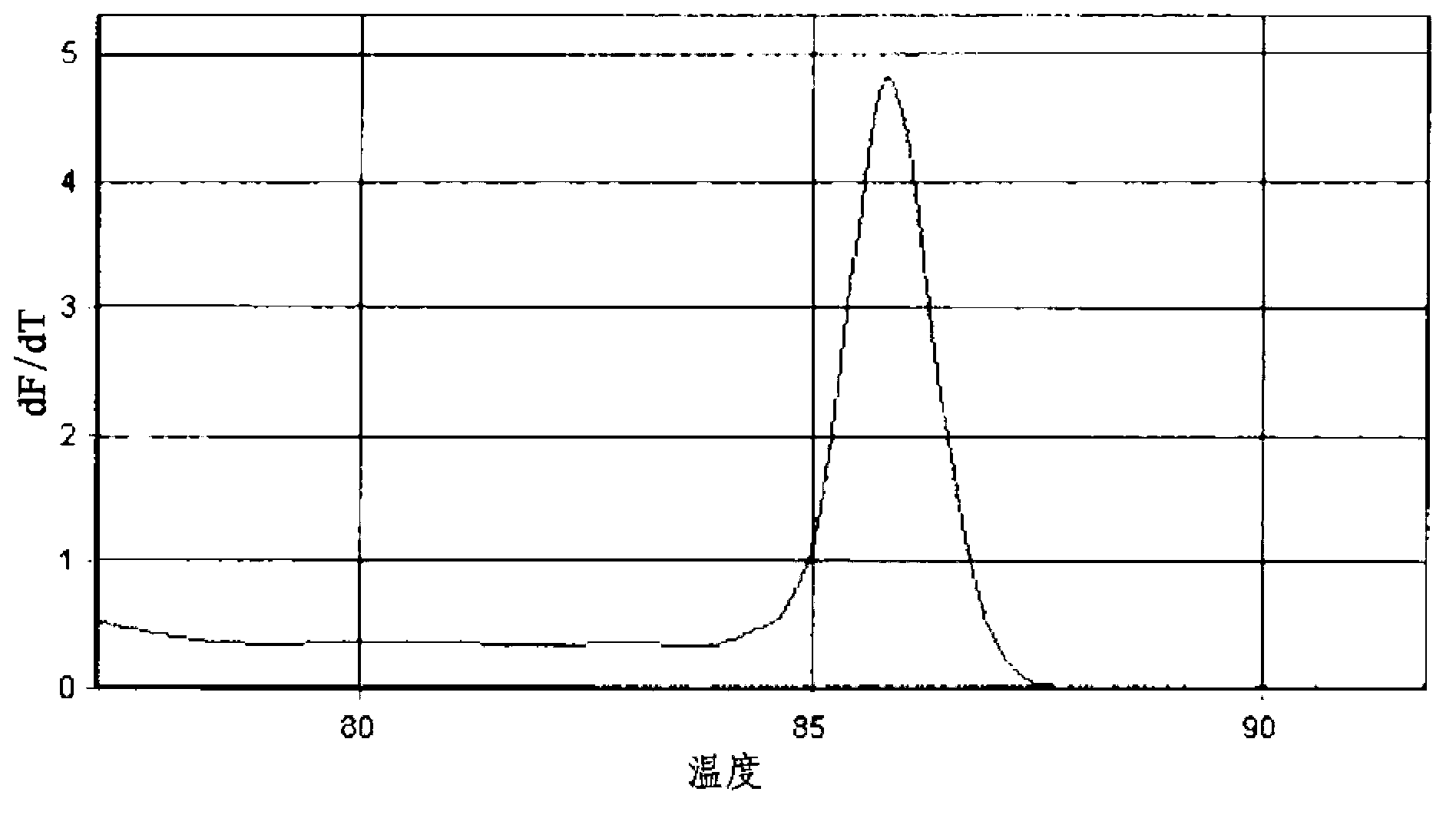
[0080] 3) PCR product size: 135bp
[0081] 4) Average melting temperature (Tm) of PCR products: about 86...
Embodiment 1-1
[0091]
[0092] (1) DNA separation
[0093] DNA was isolated from 186 mycobacterial species and 78 nontuberculous mycobacterial species, all identified in clinical subjects, and 7 standard ATCC mycobacterial species, including Mycobacterium tuberculosis (ATCC 25177) , Mycobacterium intracellulare (ATCC 13950), Mycobacterium scrofula (ATCC 19981), Mycobacterium kansasii (ATCC 12478), Mycobacterium fortuitum (ATCC 6841), Mycobacterium abscessus (ATCC 19977) and Mycobacterium avium bacilli (ATCC 25291)).
[0094] Species identified in clinical subjects (including 186 mycobacterial species and 78 nontuberculous mycobacterial species) in liquid medium (MGIT Mycobacterium medium) or solid medium (Ogawa medium) tested, or isolated directly from a sputum sample. ATCC standard species were grown in broth.
[0095] DNA was isolated from mycobacteria grown in broth as follows. Transfer 500 μL of MGIT broth cultured with mycobacteria into a 1.5 mL tube and centrifuge at 14,000 rpm f...
Embodiment 1-2
[0103]
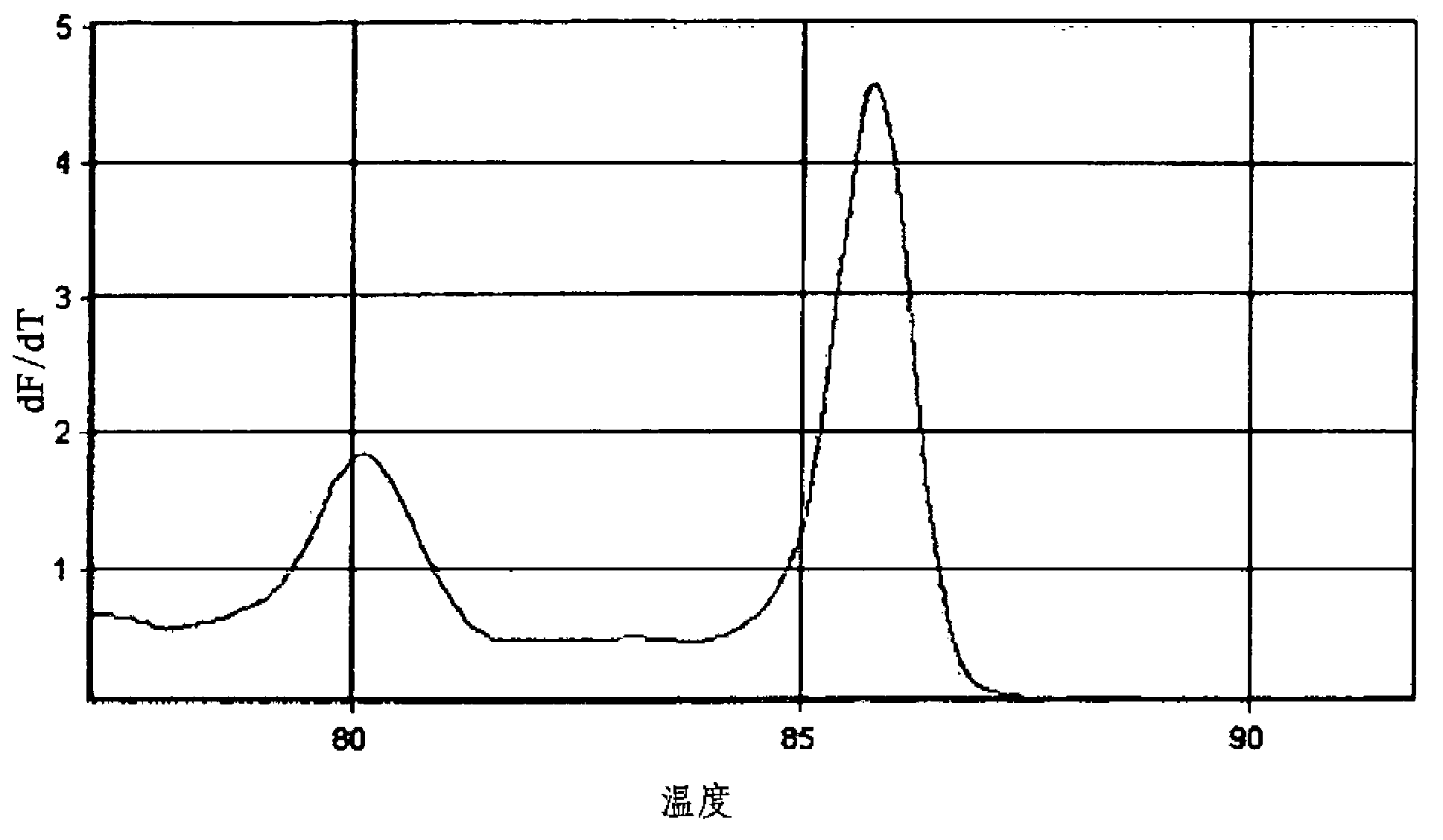
[0104] Double real-time PCR and melting curve analysis were performed in the same manner as in Example 1-1, except that the nucleotide sequence of SEQ ID NO:5 was used as the reverse primer NTM-1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com