Predictive analysis method of micro ribonucleic acid (miRNA) target genes of cattle
A technology of target gene prediction and predictive analysis, applied in special data processing applications, instruments, electronic digital data processing, etc., can solve the problem of limited number of target gene miRNA target genes, it is difficult to achieve high-throughput and large-scale, candidate gene identification Problems such as cumbersome steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
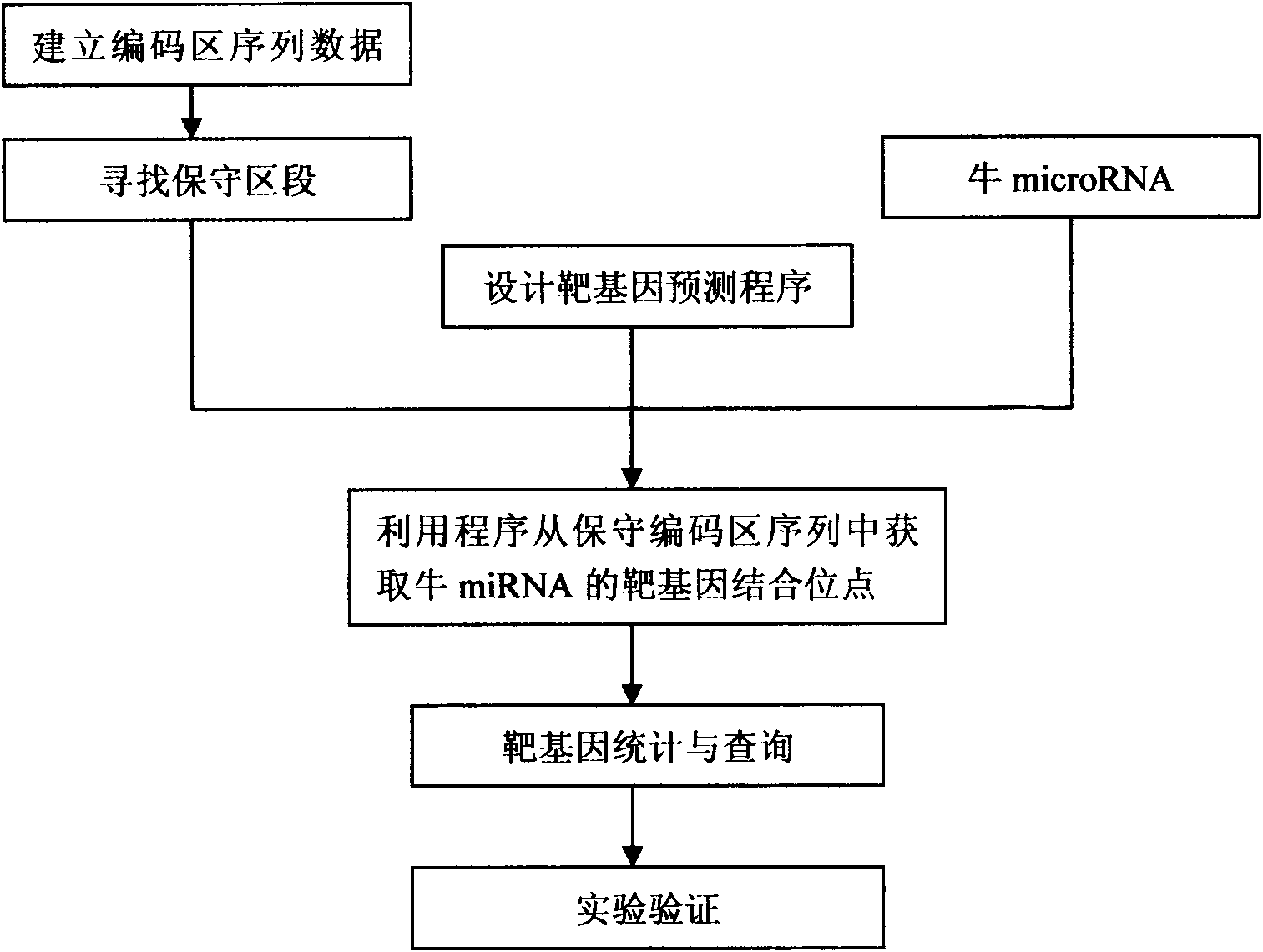
[0014] We take bovine miRNA target gene prediction as an example to illustrate the implementation process of the method of the present invention:
[0015] (1) Establish a coding region sequence database
[0016] Bovine mRNA sequences were downloaded from http: / / www.ncbi.nlm.nih.gov. By writing a PERL script program to obtain the coding protein sequence from the mRNA sequence, a coding region sequence database was established.
[0017] (2) Align the sequences of coding regions in mammals to find conserved segments.
[0018] Using the online BLAST program (http: / / www.ncbi.nlm.nih.gov / blast) to compare the sequences of mammalian coding regions, remove the poorly conserved coding region sequences, and proceed to the next step.
[0019] (3) Obtain bovine miRNA sequence information
[0020] All known mature bovine miRNA sequences were downloaded from http: / / microrna.sanger.ac.uk / sequences / .
[0021] (4) Prediction of target genes
[0022] For each microRNA, the coding regions o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com