Construction method of stably expressing clinical lamivudine-resistant hepatitis B virus cell line
A hepatitis B virus and lamivudine technology, applied in the field of stably expressing clinical lamivudine-resistant hepatitis B virus cell lines, can solve the problem of inability to truly reflect biological characteristics and differences in drug sensitivity in vitro And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
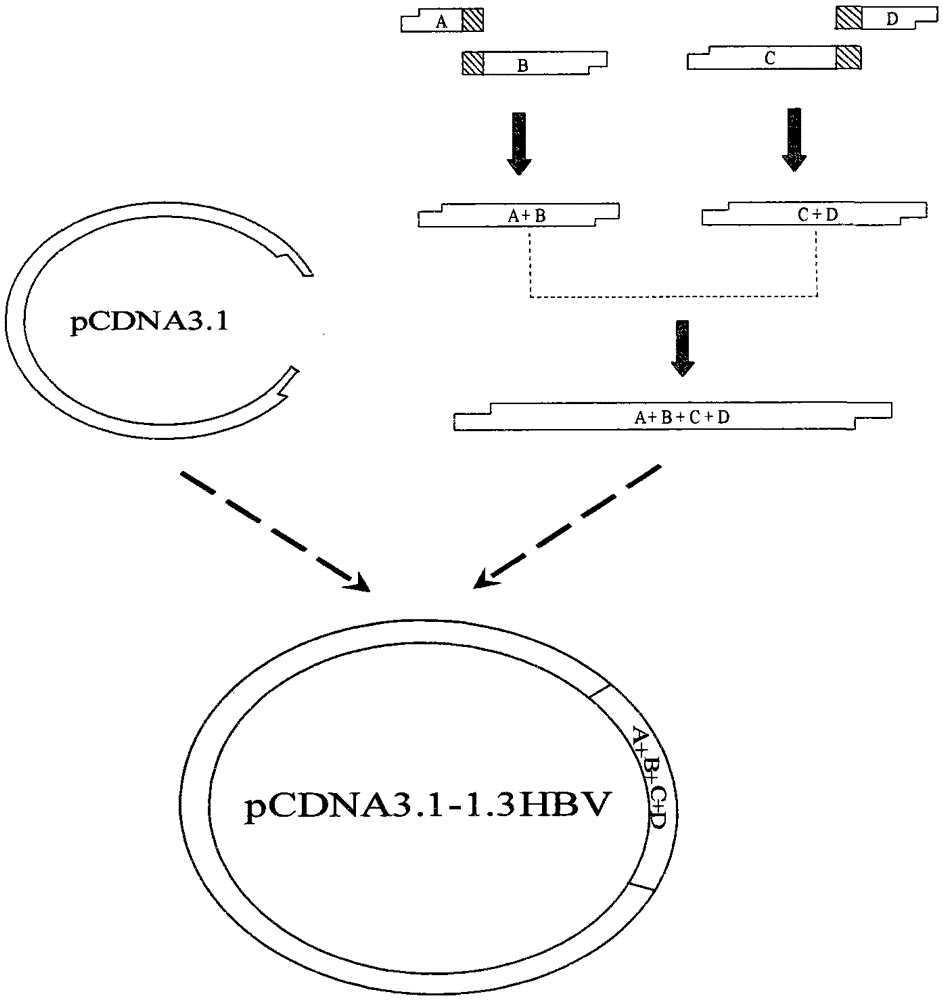
[0047] The construction of embodiment 1 HBV whole gene eukaryotic expression vector pcDNA3.1-1.3HBV
[0048] 1. Design and synthesis of primers: According to the complete HBV gene sequence in GenBank, the primers required for PCR were designed (Table 1). P1 and P2 are the upstream and downstream primers for amplifying the whole HBV gene. A1 and A2 are the upstream and downstream primers for amplifying fragment A, B1 and B2 are the upstream and downstream primers for amplifying fragment B, C1 and C2 are the upstream and downstream primers for amplifying fragment C, and D1 and D2 are the upstream and downstream primers for amplifying fragment D primers. The primer sequences are shown in Table 1 (the underlined part is the corresponding enzyme cutting site).
[0049] Table 1 PCR amplification primers for different gene fragments of HBV
[0050]
[0051] 2. PCR amplification of fragments A, B, C, D and A+B, C+D Using T-HBV containing the rtL180M+rtM204V double mutant HBV gen...
Embodiment 2
[0054] The screening of embodiment 2 stable cell lines
[0055] 1. Determination of the optimal screening concentration of G418: adjust the density of HepG2 cells to 1000 cells / ml, inoculate 1 ml per well into a 24-well culture plate. After 24 hours, add DMEM selection medium containing 100 μg / ml to 1000 μg / ml G418, change the medium every 3 days, and screen for a total of 14 days. The concentration of G418 that can just kill all cells is the optimal screening concentration of the drug. Finally, 600μg / ml was determined as the optimal screening concentration.
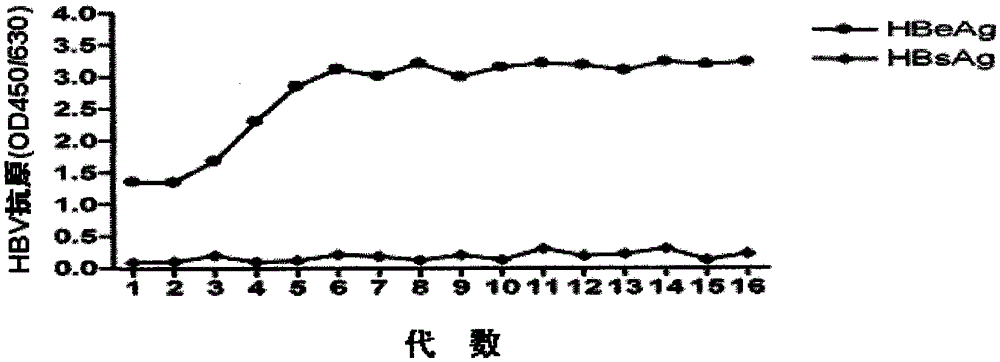
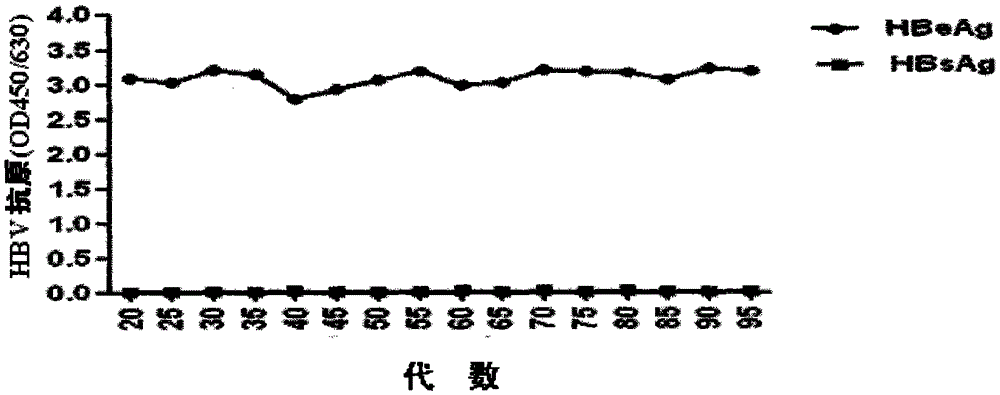
[0056] 2. Cell transfection and resistance screening Apply lipofectamion-2000 transfection reagent from Invitrogen Company, and transfect human with eukaryotic expression plasmid pcDNA3.1-1.3HBV containing rtL180M+rtM204V double variant without endotoxin in strict accordance with the operating instructions Liver cancer cell line HepG2 cells. 24 h after transfection, the transfected cells were passaged to a new culture ...
Embodiment 4
[0062] The drug resistance identification of embodiment 4 stable cell lines
[0063] 1. Identification of phenotypic drug resistance Taking the currently recognized HepG2.2.15 cell line stably expressing wild-type HBV as a control, evaluate the stable cell line HepG2.RL1 established by the present invention against lamivudine and adefovir dipivoxil in vitro sensitivity. Adjust the cell concentration of HepG2.2.15 and HepG2.RL1 to 2×10 4 cells / ml, 200 μl per well was inoculated into a 96-well cell culture plate. At the same time adjust the cell concentration to 2×10 5 cells / ml, 2ml per well was inoculated into a 96-well cell culture plate. After culturing for 24 hours, culture solutions containing different concentrations of lamivudine or adefovir dipivoxil (0, 0.01, 0.1, 1, 10, 100 μM) were added, and three replicate wells were set for each concentration in a 96-well plate. Two replicate wells were set up for each concentration in the 6-well plate. Cell culture medium was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com