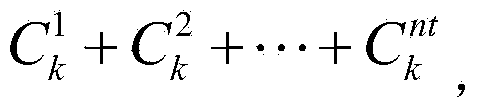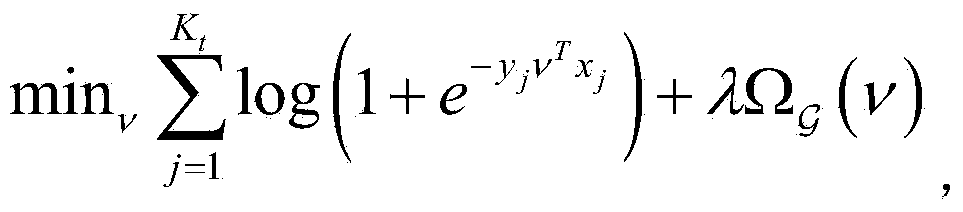Gene sequence sorting method based on combination map rarefaction
A technology of gene sequence and classification method, applied in the field of computer biological information processing, can solve problems such as difficulty in use, increase in feature space, and inability to use computers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Assuming a gene sequence classification problem, the gene sequence to be classified is:
[0079] A. Positive class: AAGA, denoted as d 1
[0080] B. Negative class: ATTG, denoted as d 2
[0081] If represented by a first-order template, the feature space becomes: A, C, T, G, A, C, T, G, A, C, T, G, A, C, T, G. The first four features represent the four possibilities corresponding to position 1, the 5-8 features represent the four possibilities corresponding to position 2, the 9-12 features represent the four possibilities corresponding to position 3, and the 13-16 features represent the four possibilities corresponding to position 4 Corresponding four possibilities. According to the vector representation method described above, it is finally expressed in the form of Table 1:
[0082] Table 1
[0083] category
Gene sequence vector representation
positive class
x 1 =(1,0,0,0,1,0,0,0,0,0,0,1,1,0,0,0)
negative class
x 2 =(1,0,0,0,0,0...
Embodiment 2
[0107] Algorithms used in the present invention are all written and realized by python language. The model used in the experiment is: Intel Xeon X7550 processor, the main frequency is 2.00G HZ, and the memory is 32G. The SPAMS toolkit used in the present invention is a general open source classifier training package at present.
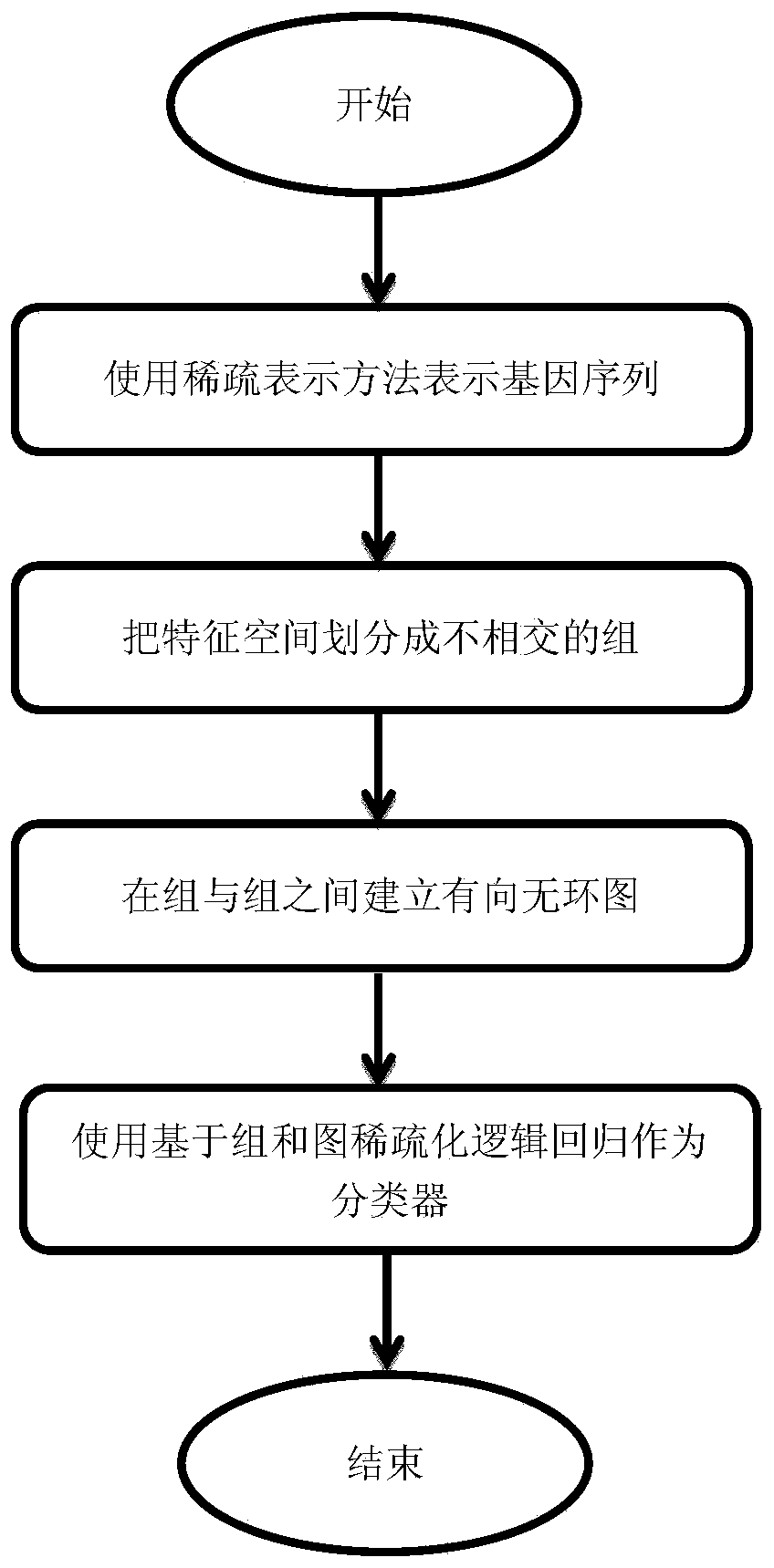
[0108] More specifically, as figure 1 As shown, the present invention operates as follows:
[0109] 1. Group the feature space: use sparse representation to express each gene sequence as a vector, and divide the entire feature space into mutually disjoint groups. The feature space is established using the first-order, second-order, and third-order templates, and the grouping is also grouped according to the first-order, second-order, and third-order templates;
[0110] 2. Establish a directed acyclic graph between groups: establish a directed acyclic graph between groups, and assign a cost value (cost) to each edge on the graph;
[0111] 3. Classi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com