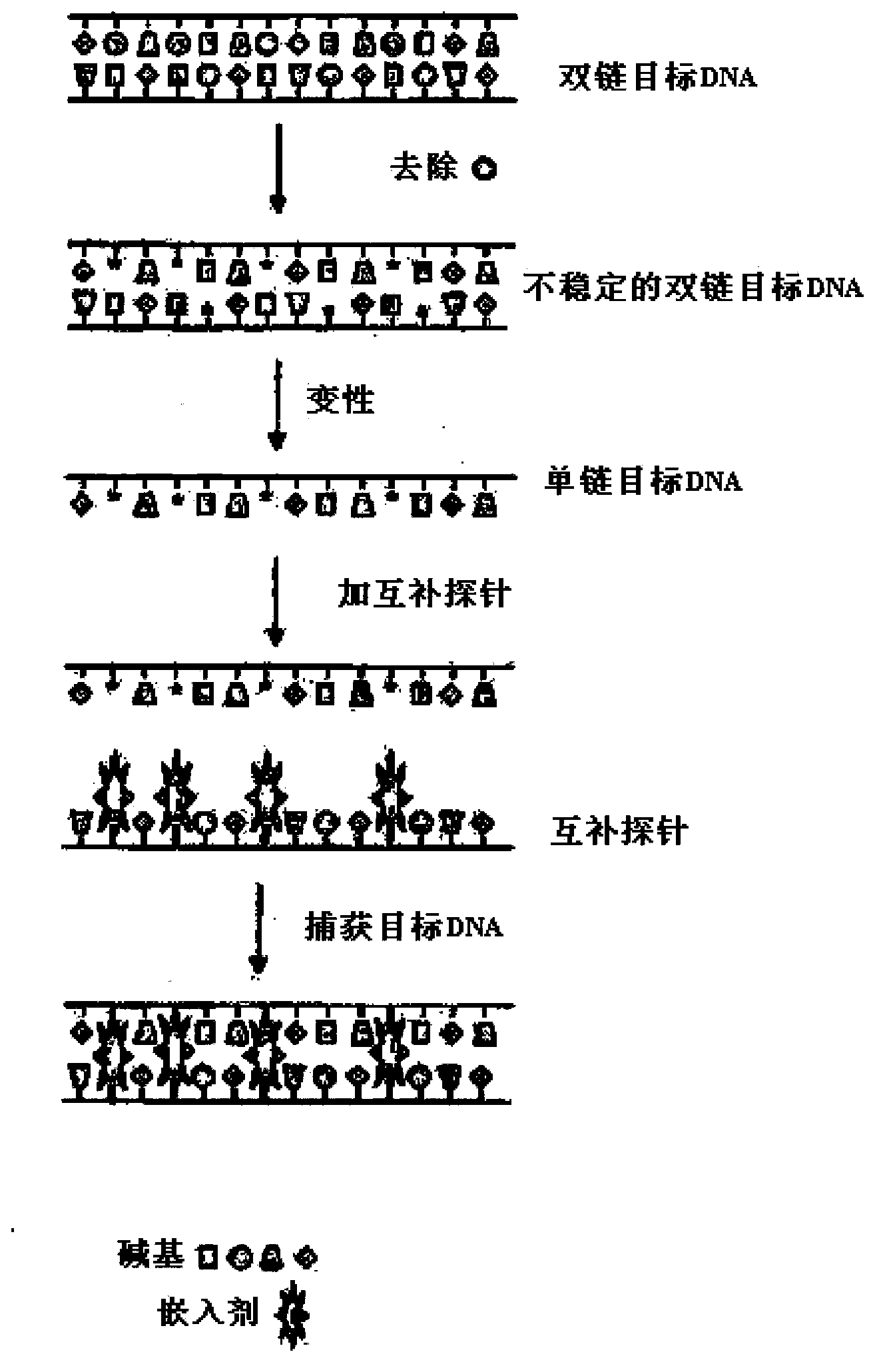
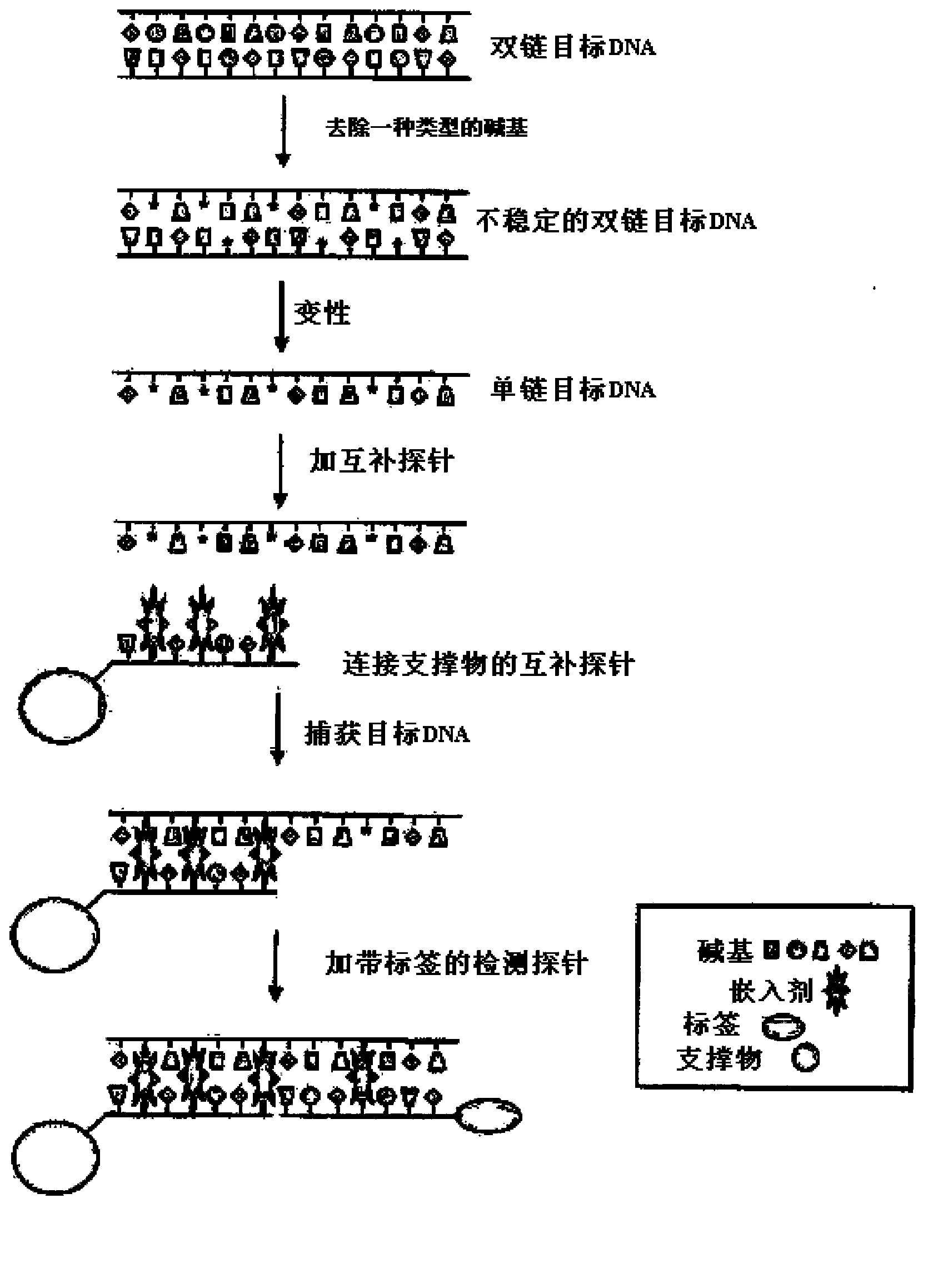
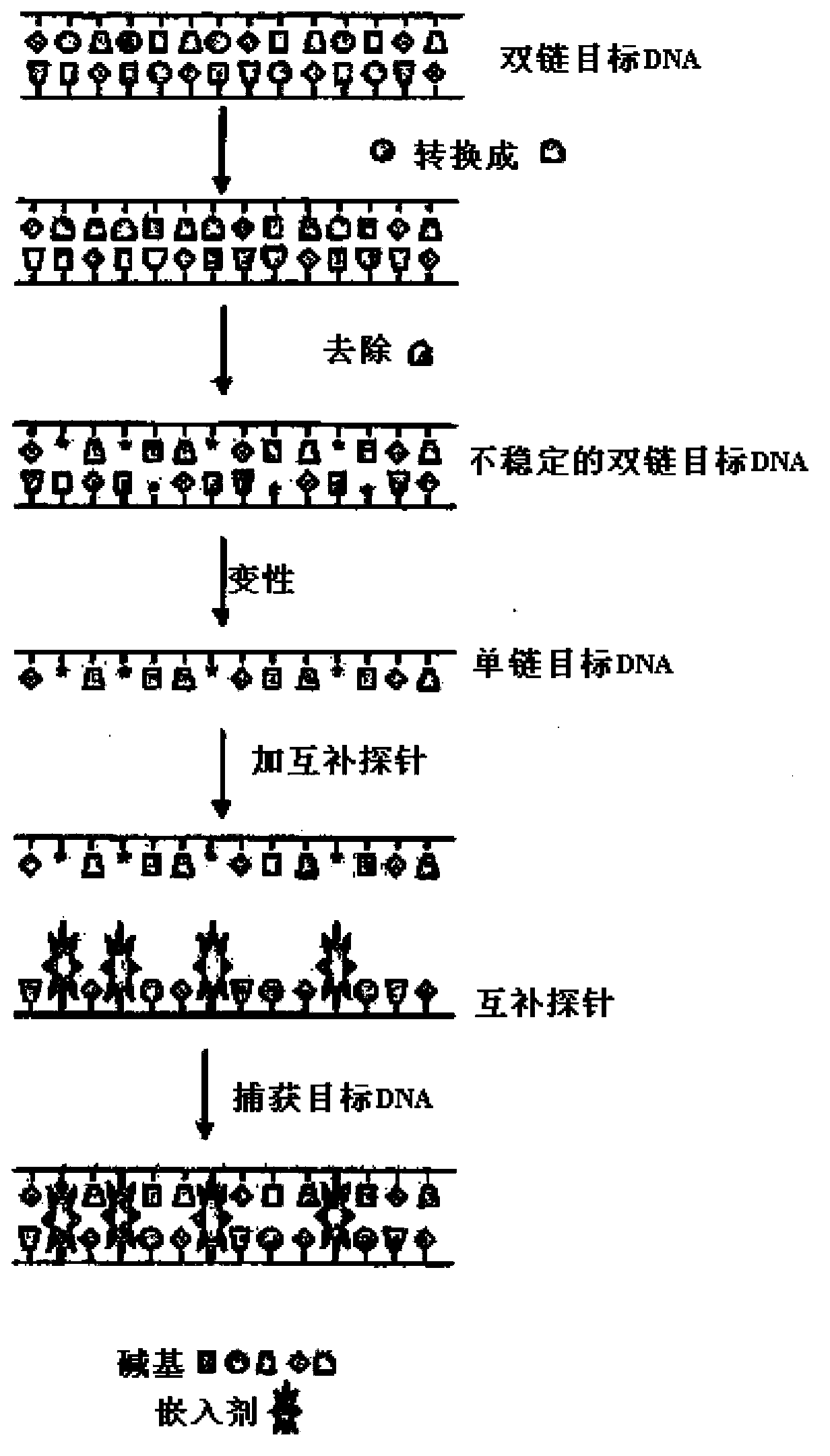
Capture of target DNA and RNA by probes comprising intercalator molecules
An intercalator, target technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as insufficient identification of base mismatches
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0517] Assessing the effect of abasic sites on Tm by obtaining melting curves
[0518] use A Fluorescence Resonance Energy Transfer (FRET) system on 2.0 evaluates changes in melting point (Tm) caused by base mismatch or debasic sites (B) in two different oligonucleotide sequences.
[0519] Oligonucleotides were purchased from IBM GmbH ( Germany) or DNA Technology A / S (Risskov, Denmark), high performance liquid chromatography (HPLC) purification and subsequent quality control were performed on a 0.2 μmol synthesis scale. Oligonucleotides were synthesized using 3' amino-modifier-C7 followed by attachment to ATTO495NHS-ester, or using 5' amino-modifier-C6 followed by attachment to ATTO590NHS-ester. ATTO495 acts as a FRET donor and is a modification of acridine orange with an excitation maximum at 495nm and an emission maximum at 527nm. ATTO590 is a derivative of Rhodamine dye with an excitation maximum at 594nm and an emission maximum at 624nm. Pair ATTO495 / ATTO590FRET in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com