Specific PCR (Polymerase Chain Reaction) identification method of paecilomyces hepiali powder
A technology of Paecilomyces batmoth and Paecilomyces batmoth, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and determination/inspection of microorganisms, which can solve the limitation of industrial upgrading and standardized development of Paecilomyces batmoth , product quality is difficult to guarantee and other problems, to achieve the effect of low cost, high accuracy and strong primer specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
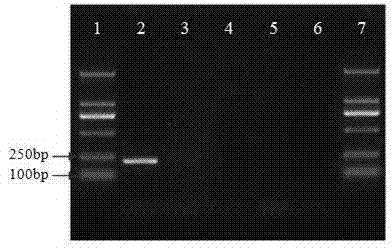
[0025] Example 1: Design of PCR primers and detection of samples
[0026] 1 Paecilomyces bat moth β-tubulin Design of gene-specific PCR primers
[0027] According to the publicly registered Paecilomyces bat moth on GenBank β-tubulin Gene sequence information for primer design (see attached figure 1 ).
[0028] The GenBank accession number of the reference sequence is: AY624217.
[0029] An upstream primer was designed at 18-35 and named as CICC-Pae-F1. The sequence is: 5'-GAGCATGGT CTCGACTCT-3'.
[0030] A downstream primer was designed at 233-249 and named as CICC-Pae-R1. The sequence is: 5'-CGAAGGG ACCAGCACGG-3'.
[0031] 2 Detection of samples
[0032] 2.1 Collection of samples
[0033] Buy commercially available Paecilomyces bat moth powder, Cordyceps sinensis fungus powder, Cordyceps sinensis, and Cordyceps militaris fungus powder.
[0034] 2.2 Sample genomic DNA extraction
[0035] Genomic DNA was extracted by the modified CTAB method and purifie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com