T-hairpin structure-mediated method for measuring unknown sequence of DNA flank
A DNA sequence, unknown sequence technology, applied in the field of bioengineering, can solve the problems of single connection site, low linker connection efficiency, high price, etc., and achieves the effect of wide application range, labor saving and low cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
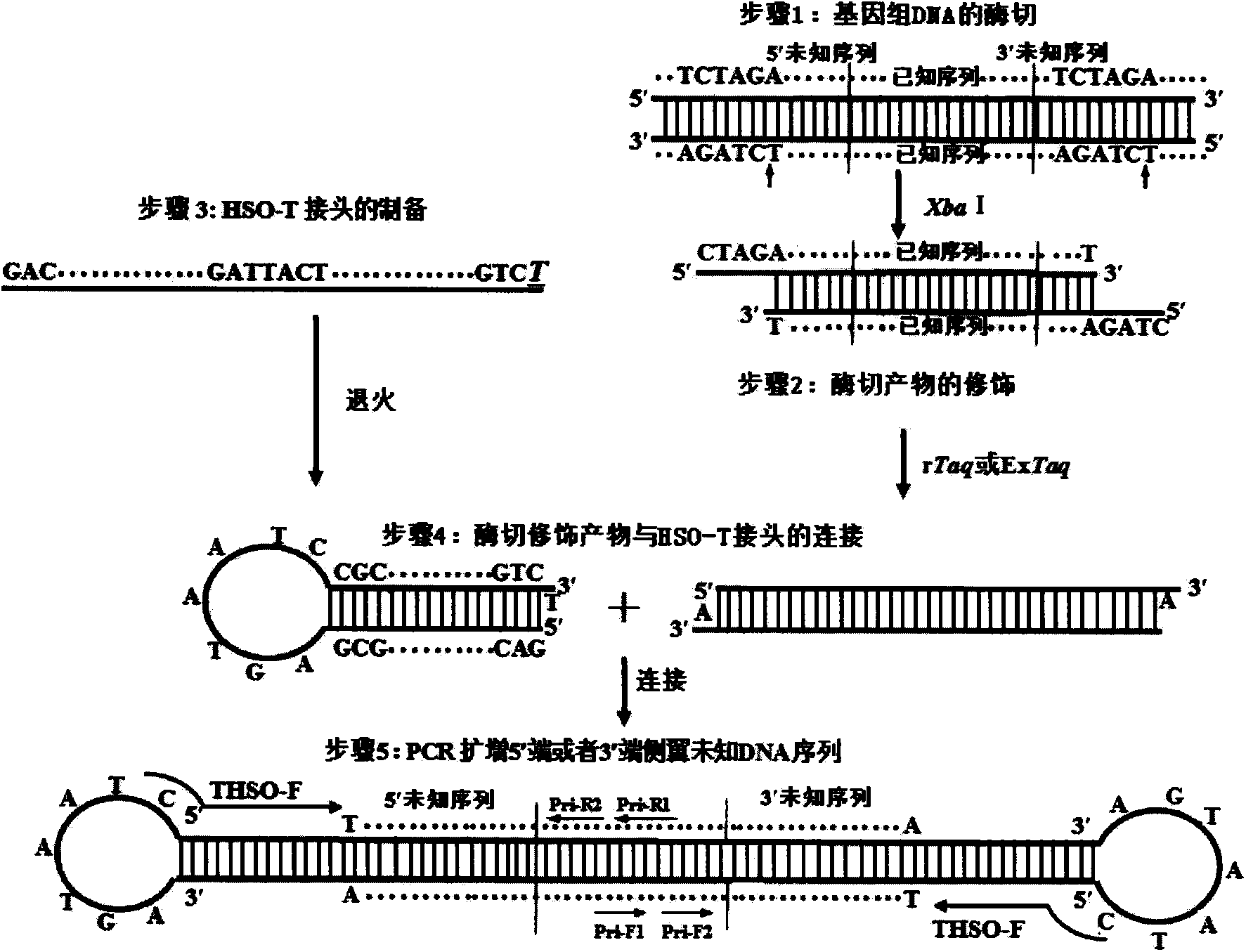
[0029] Aspergillus usamii (Aspergillus usamii) E001Auseh2 gene 5' flanking unknown DNA sequence determination:
[0030] (1) Construction of HSO-T linker: HSO-T (100 μmol / L) 2 μL, ddH 2 O8 μL, mix well; denature with a PCR gene amplification instrument at 94°C for 3 minutes, slowly cool down (1°C / s) to 25°C, and then anneal at a constant temperature for 30 minutes.
[0031] (2) Preparation of genomic DNA digested products: Xba I was used to digest the genomic DNA of Aspergillus usami. Construct the following digestion system: 10×M Buffer 1 μL, Xba I 0.5 μL, 0.1M BSA 1 μL, genomic DNA 5 μL, ddH 2 O2.5μL; react at 37°C for 4h.
[0032] (3) Enzyme digestion genome modification: 20 μL of enzyme digestion product, 2.5 μL of 10×PCR Buffer, 0.5 μL of dNTP, 0.25 μL of rTaq or Ex Taq, ddH 2O1.75μL; use a PCR gene amplification instrument to react at 72°C for 10min; the modified enzyme-digested product is named Auseh2-DM.
[0033] (4) Connection of Auseh2-DM and HSO-T adapter: 10×T4 ...
Embodiment 2
[0040] Determination of the unknown DNA sequence flanking the 3' end of the Aspergillus usamii E001Auseh2 gene:
[0041] (1) The preparation and connection method of the HSO-T linker and the genomic DNA digestion product are the same as in Example 1.
[0042] (2) Synthesis of upstream primers with known sequences: based on the known Auseh2 gene sequence, two specific downstream primers were synthesized to amplify the unknown sequence flanking the 3′ end of the known DNA sequence, named EH-F1 and EH-F2, Where EH-F2 is more than 30bp away from the 3′ end of the known DNA sequence:
[0043] EH-F1: 5′-TCATGATGCTTGCAAAGCC-3′
[0044] EH-F2: 5′-GGAGAGAAGTTCCTCGAT-3′
[0045] (3) DNA sequence amplification at the 3′ end of Auseh2: 10×PCR Buffer 2.5 μL, dNTP 1.5 μL, THSO-PCR template 2 μL, THSO-F 0.5 μL, EH-F 10.5 μL, ddH 2 O17.75 μL, rTaq enzyme 0.25 μL; 94°C for 5 min, 30 cycles (94°C, 30s; 51°C, 30s; 72°C, 1min20s), 72°C for 10min, the first round of PCR amplification product wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com