T vector mediated method for testing 3' end flanking unknown sequence
A technology of unknown sequence and carrier, applied in the field of bioengineering, can solve the problems of not being able to determine the sequence of the upstream and downstream regulatory elements of the gene, not being able to study the structure and function in depth, and consuming a lot of manpower and material resources, so as to save manpower, operate with fewer restrictions, low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
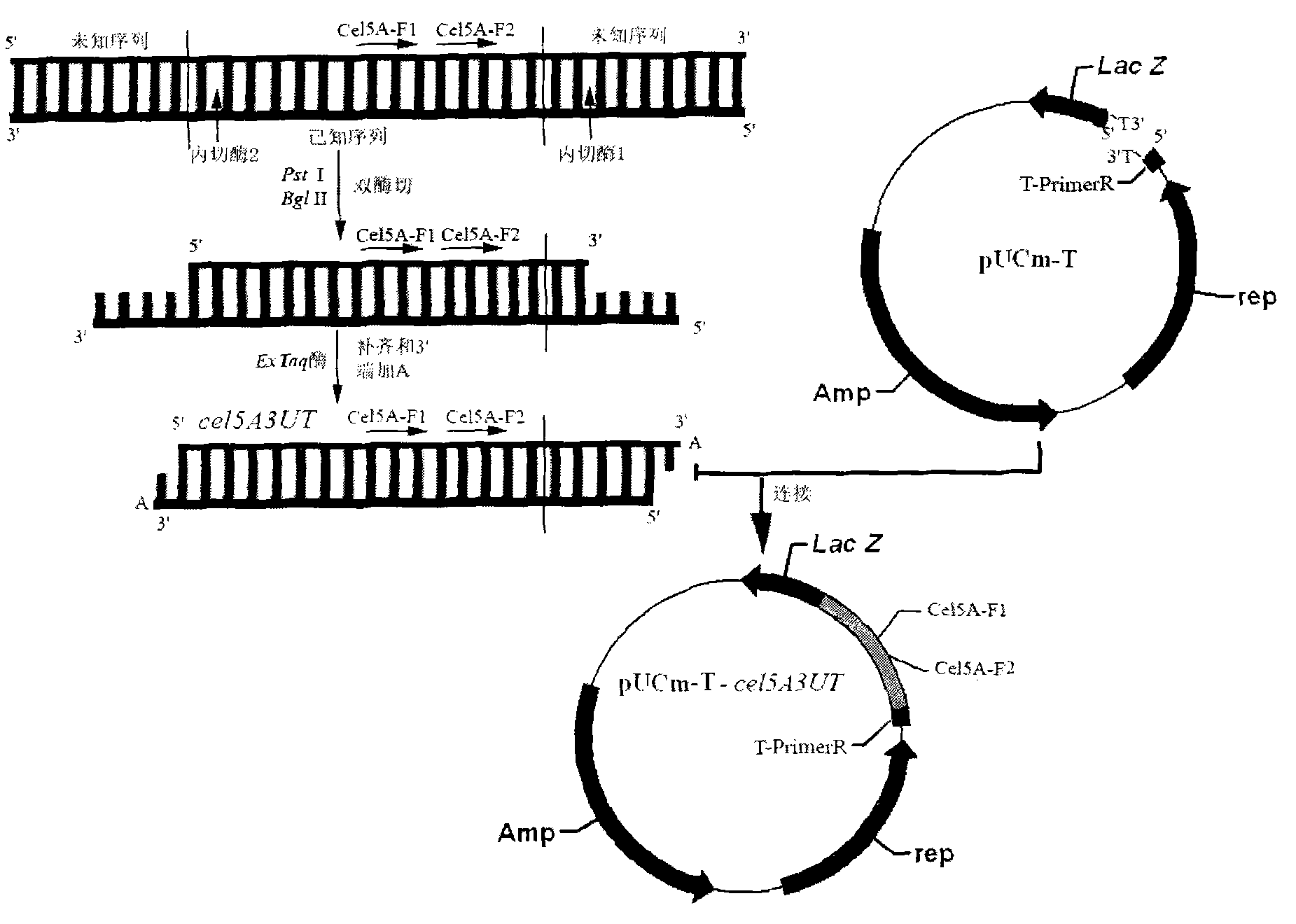
[0028] Determination of the 3' flanking sequence of Aspergillus usamii E001 cel5A.
[0029] (1) Preparation of DNA double-enzyme digestion products: PstⅠ and BglII were used to perform double-digestion of Aspergillus usami genomic DNA. The following double enzyme digestion system was constructed: 10×H Buffer 2 μl, PstⅠ 1 μl, BglⅡ 1 μl, genomic DNA 10 μl, sterile water 6 μl; react the system in a 37°C water bath for 4 hours. Purify and recover the double-digested product and dissolve it in 20 μl sterile water.
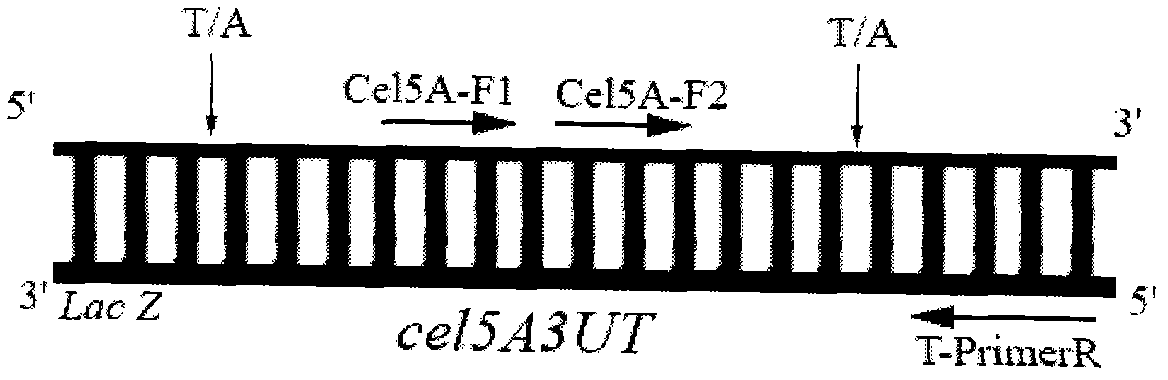
[0030] (2) Complementation of digested products and addition of A at the 3′ end: 20 μl of double digested products, 2.5 μl of 10×Ex Taq Buffer, 0.5 μl of dNTP, 0.25 μl of Ex Taq enzyme, 1.75 μl of sterile water; 72°C reaction 10min. The target product was named cel5A3UT.
[0031] (3) Ligation of cel5A3UT and pUCm-T: 1 μl of 50% PEG4000, 1 μl of 10×T4 DNA Ligase Buffer, 1 μl of pUCm-T, 1 μl of T4 DNA Ligase, 6 μl of cel5A3UT; overnight at 16°C. The target product was...
Embodiment 2
[0036] Determination of the 3' flanking sequence of Aspergillus usamii E001 cell2A.
[0037] (1) Preparation of DNA digestion products: SalI and BamHI were used to perform double enzyme digestion on the genomic DNA of Aspergillus usami. The following enzyme digestion system was constructed: 3 μl of 10×T Buffer, 1 μl of SalⅠ, 1 μl of BamHI, 10 μl of genomic DNA, and 5 μl of sterile water; the system was reacted in a water bath at 37°C for 4 hours. Purify and recover the double-digested product and dissolve it in 20 μl sterile water.
[0038] (2) Completion of digested products and addition of A at the 3′ end: 20 μl of double digested products, 2.5 μl of 10×PCR Buffer, 0.5 μl of dNTP, 0.25 μl of Taq enzyme, 1.75 μl of sterile water; react at 72°C for 10 minutes. The target product was named cel12A3UT.
[0039] (3) Ligation of cel12A3UT and pUCm-T: 1 μl of 50% PEG4000, 1 μl of 10×T4 DNA Ligase Buffer, 1 μl of pUCm-T, 1 μl of T4 DNA Ligase, 6 μl of cel12A3UT; overnight at 16°C. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com