Hairpin structure-mediated method for determining unknown sequence of 3'-end flanking region
A hairpin structure and unknown sequence technology, applied in the field of bioengineering, can solve the problems of uncertain gene upstream and downstream regulatory element sequences, low reverse transcription abundance, high-level structure, mRNA instability, etc., to save manpower, less operation restrictions, low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
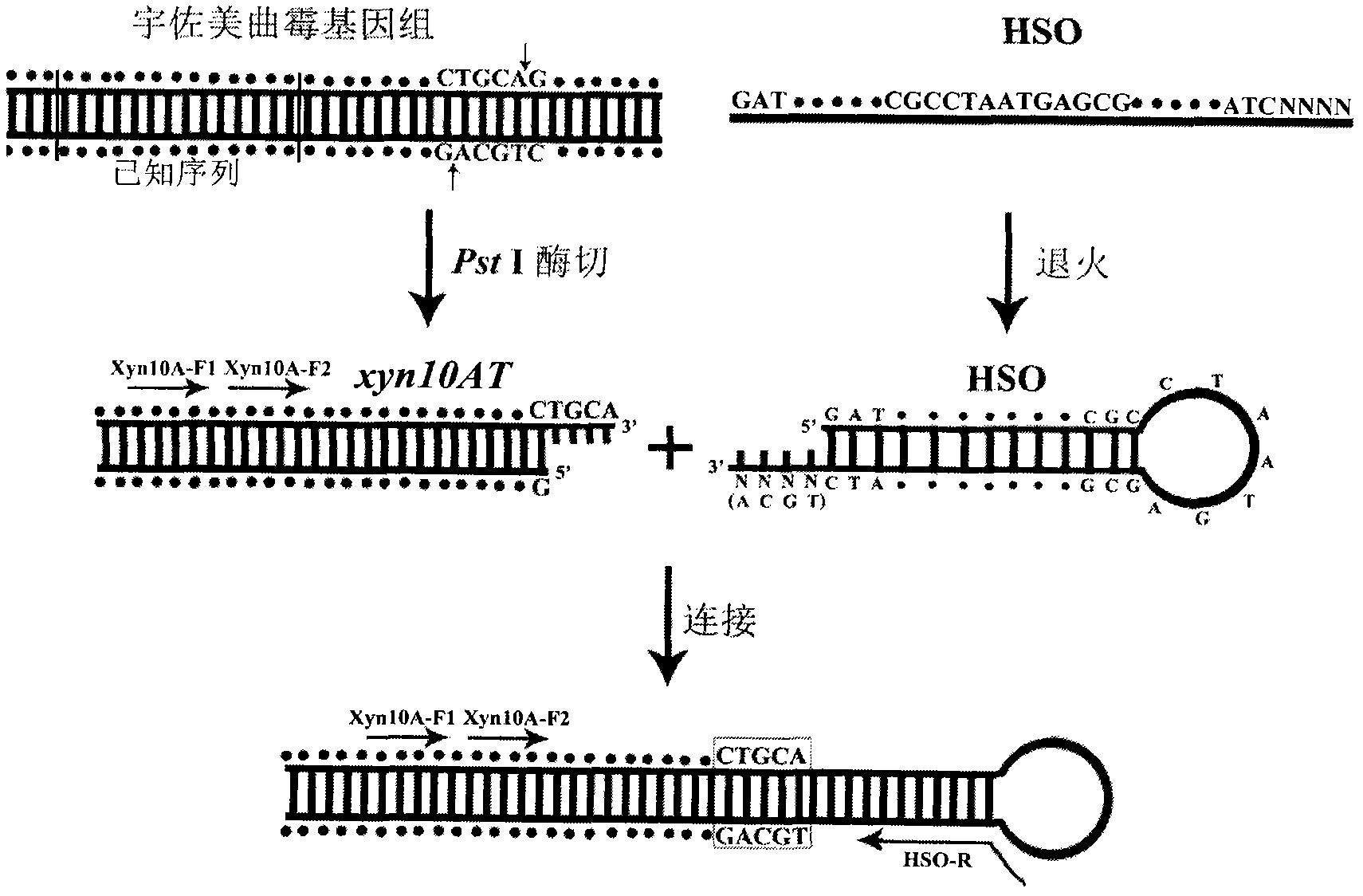
[0030] Sequence determination of the 3' regulatory region of Aspergillus usamii E001 xyn10A.
[0031] (1) Preparation of DNA digestion products: PstI was selected for single digestion of Aspergillus usami genomic DNA. The following enzyme digestion system was constructed: 10×H Buffer 2 μl, PstI 2 μl, genomic DNA 10 μl, sterile water 6 μl; the above system was reacted in a 37°C water bath for 4 hours. The purified product (20 μl) was named xyn10AT.
[0032] (2) Preparation of the hairpin structure: HSO (100 μmol / L) 2 μl, sterile water 8 μl, mix well; denature at 94°C for 3 minutes, slowly cool down (1°C / s) to 25°C and then anneal at constant temperature for 30 minutes.
[0033] (3) Ligation of xyn10AT and hairpin structure: 1 μl of 10×T4 DNA Ligase Buffer, 3 μl of HSO, 5 μl of xyn10AT, 1 μl of T4 DNA Ligase; overnight at 16°C.
[0034] (4) The first round of PCR for the regulatory region of xyn10A 3′ end: 10×PCR Buffer 2.5 μl, dNTP 1.5 μl, HSO linker 2 μl, Xyn10A-F10.5 μl, HS...
Embodiment 2
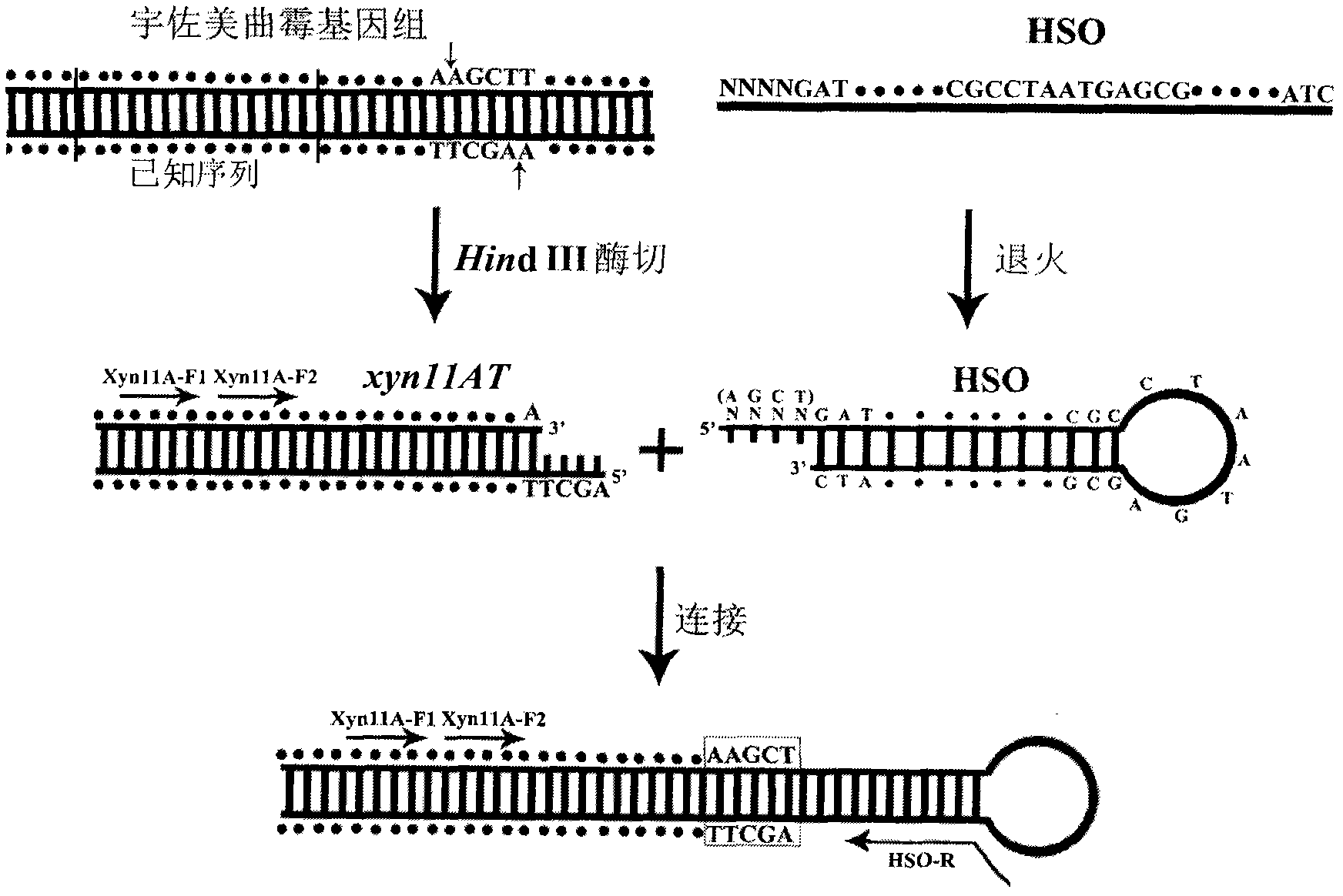
[0038] Sequence determination of the 3' regulatory region of Aspergillus usamii E001 xyn11A.
[0039] (1) Preparation of DNA digestion products: HindⅢ was used to digest the genomic DNA of Aspergillus usami. The following enzyme digestion system was constructed: 10×M Buffer 2 μl, HindⅢ 2 μl, 0.1% BSA 2 μl, genomic DNA 10 μl, sterile water 4 μl; react the system in a 37°C water bath for 4 hours. The digested and purified product (20 μl) was named xyn11AT.
[0040] (2) Preparation of hairpin joints: 2 μl of HSO (100 μmol / L), 8 μl of sterile water, fully mixed; denatured at 94°C for 3 minutes, slowly cooled (1°C / s) to 25°C and then annealed at constant temperature for 30 minutes.
[0041] (3) Ligation of xyn11AT and hairpin structure: 1 μl of 10×T4 DNA Ligase Buffer, 3 μl of HSO, 5 μl of xyn11AT, 1 μl of T4 DNA Ligase; overnight at 16°C.
[0042] (4) The first round of PCR of xyn11A 3′-end regulatory region: 10×PCR Buffer 2.5μl, dNTP 1.5μl, HSO linker 2μl, Xyn11A-F10.5μl, HSO-R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com