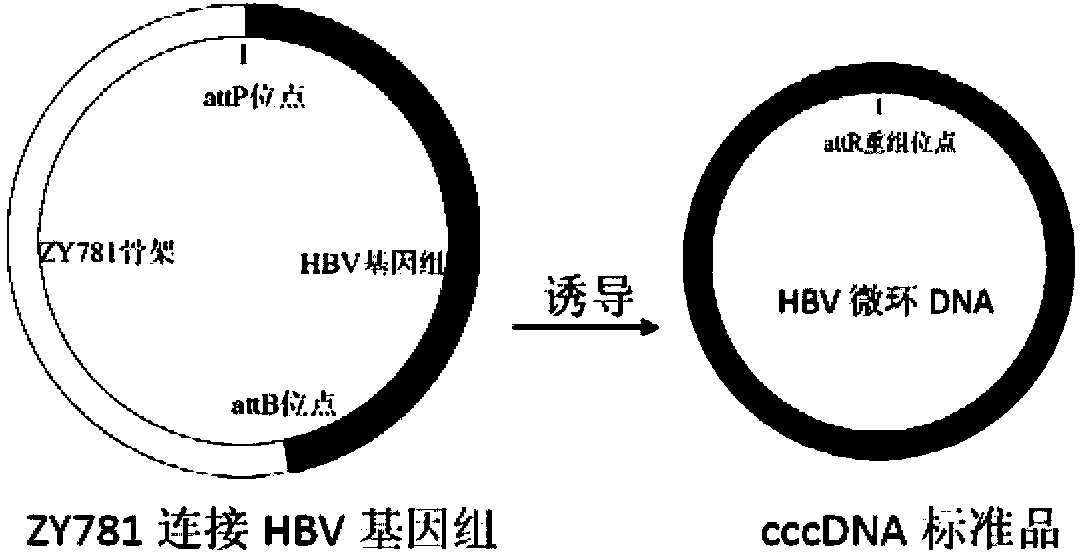
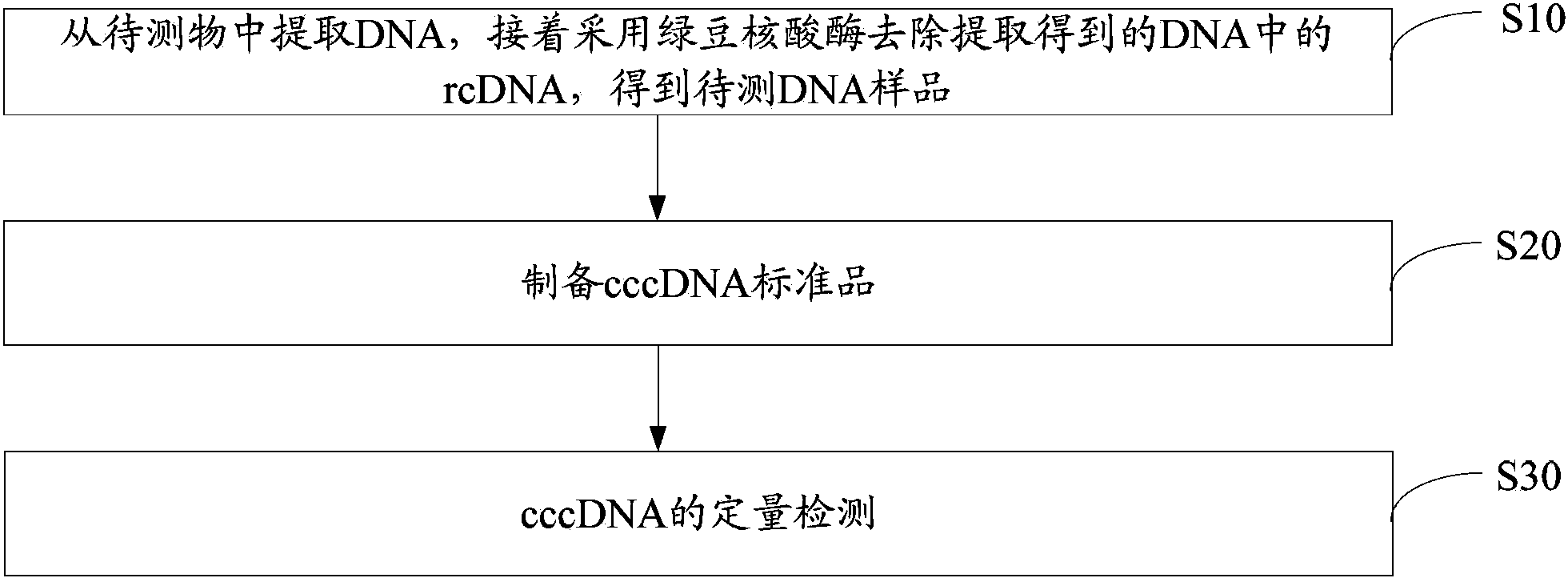
cccDNA standard substance, preparation method thereof, and method and kit for quantitatively detecting cccDNA of hepatitis b virus
A technology for quantitative detection of hepatitis B virus, applied in the field of molecular biology, can solve problems such as difficulty in obtaining cccDNA, inaccurate quantification of cccDNA, and impact on the process and results of fluorescent quantitative PCR
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
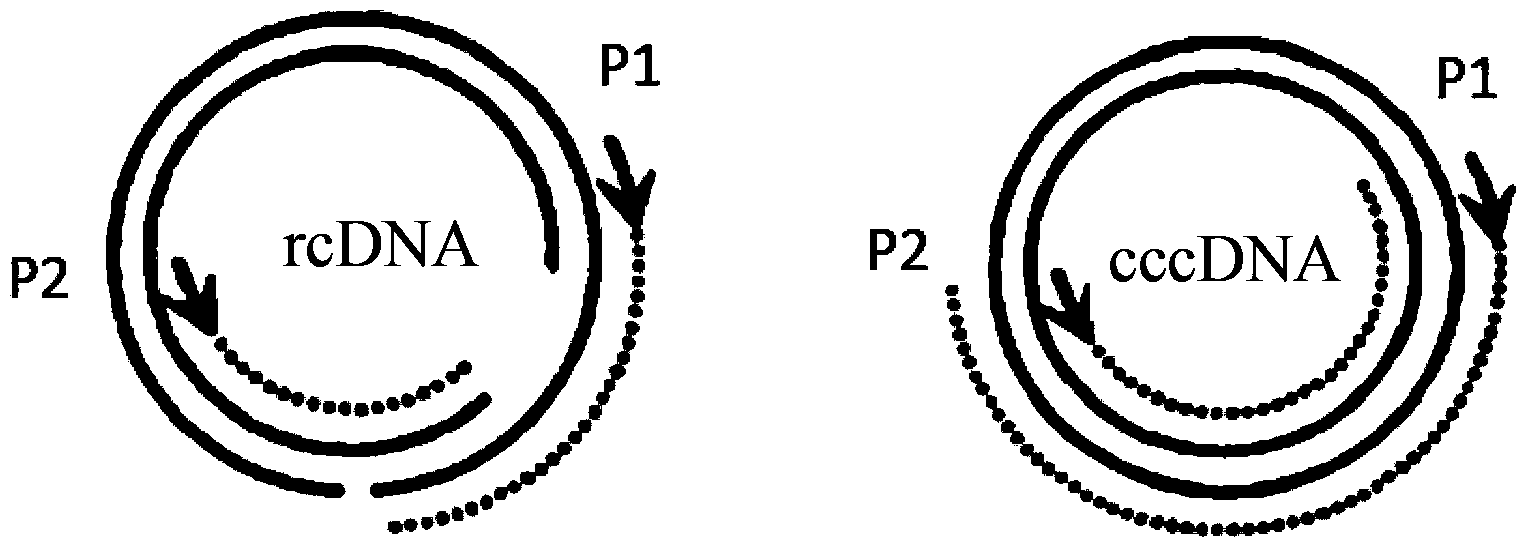
[0036] Such as figure 1 As shown, the traditional detection of cccDNA by real-time fluorescent quantitative PCR is realized by a pair of special primers spanning the double gap of rcDNA and a fluorescent probe. The sense primer P1 is complementary to the negative strand and is located upstream of the positive strand gap. The antisense primer P2 is complementary to the positive strand and is located downstream of the negative strand gap. rcDNA is not amplified because of the gap, but cccDNA can be selectively amplified . A fluorescent probe complementary to the negative strand is added downstream of the cccDNA negative strand gap, which can realize the quantitative analysis of cccDNA by fluorescent quantitative PCR.
[0037]However, the method for detecting cccDNA by real-time fluorescent quantitative PCR has the following disadvantages: 1. When PCR detects cccDNA, due to the presence of rcDNA, the amount of initial template cannot be too high, otherwise primers P1 and P2 will...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com