New Short Nucleotide Tandem Repeat Sequence Site and Its Application
A short tandem repeat and nucleotide sequence technology, applied in the field of short nucleotide tandem repeat sequence sites, can solve problems such as poor compatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
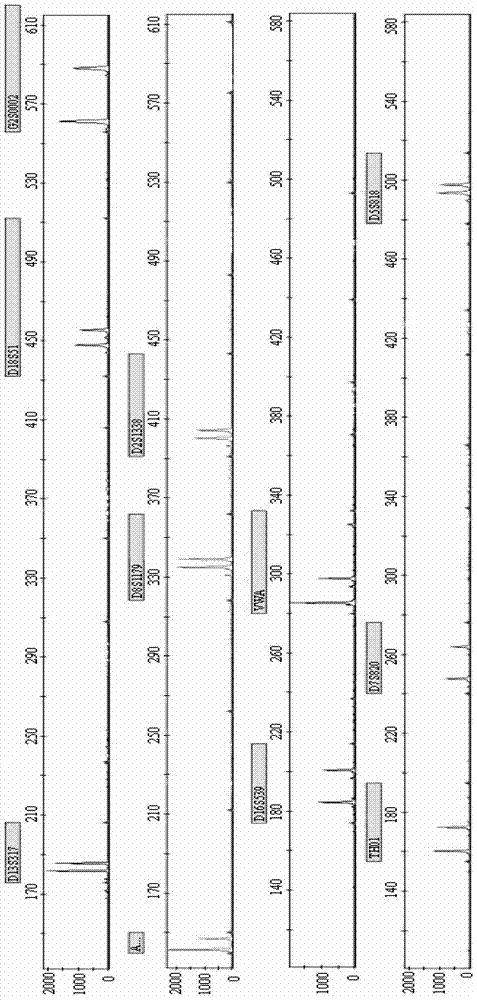
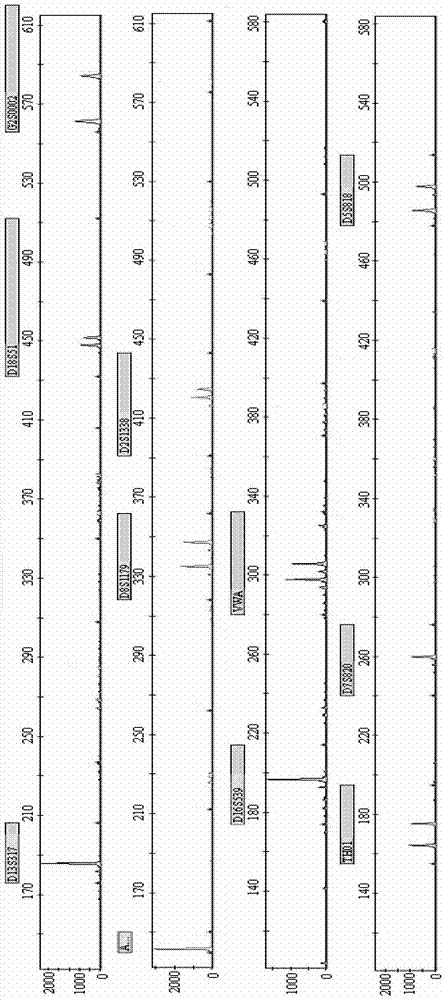
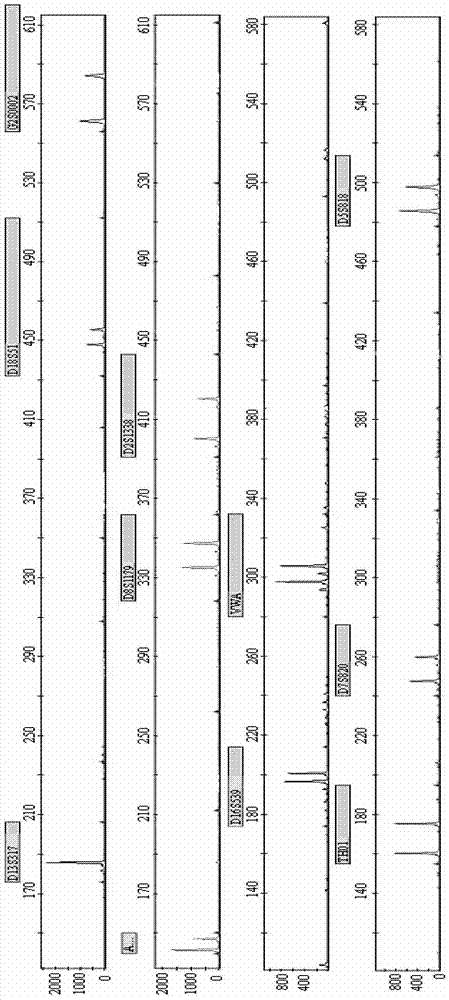
[0129] Combining the new STR locus G2S0002 of the present invention with 10 commonly used loci (VWA, D16S539, D5S818, D7S820, D13S317, D8S1179, D18S51, TH01, D2S1338, AMELO), multiplex fluorescent PCR technology was used to check the samples, and the four used The samples were three samples from a related family and one unrelated control sample. The primer sequences of G2S0002 and 10 commonly used loci are shown in Table 2.
[0130] Table 2 Primer Sequence
[0131]
[0132] Note: PET, VIC, NED, FAM four fluorescent dyes ((Applied Biosystems, USA company).
[0133] The specific experimental steps are as follows:
[0134] 1) DNA sample preparation
[0135] Collect blood from 4 selected sample individuals; extract DNA samples through DNA extraction kit and take 1 μl each, perform quality inspection and concentration estimation on the samples by 1% agarose electrophoresis, and then dilute the samples to the working concentration according to the estimated concentration 5-10...
Embodiment 2
[0147] Randomly select 20 groups of related and unrelated samples, and use the method of Example 1 for detection, the difference is that only the new STR locus G2S0002 of the present invention is used.
[0148] After testing, after determining the paternal sample, determine the offspring samples that are related to the male parent in the remaining 19 groups of samples. The new STR locus G2S0002 of the present invention can accurately exclude 12 groups of unrelated samples. A higher degree of discrimination was obtained.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com