Construction method of single enzyme digestion vector
A construction method and carrier technology, applied in the field of DNA recombination, can solve the problems of increasing the difficulty of operation steps for nucleic acid degradation, reducing the efficiency of single-enzyme digestion and connection, and long connection time, so as to improve the efficiency of single-enzyme digestion and connection, improve the efficiency of single-enzyme The effect of cutting connection efficiency and saving manpower
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 Construction of pBC1-HIOMT mammary gland-specific expression vector
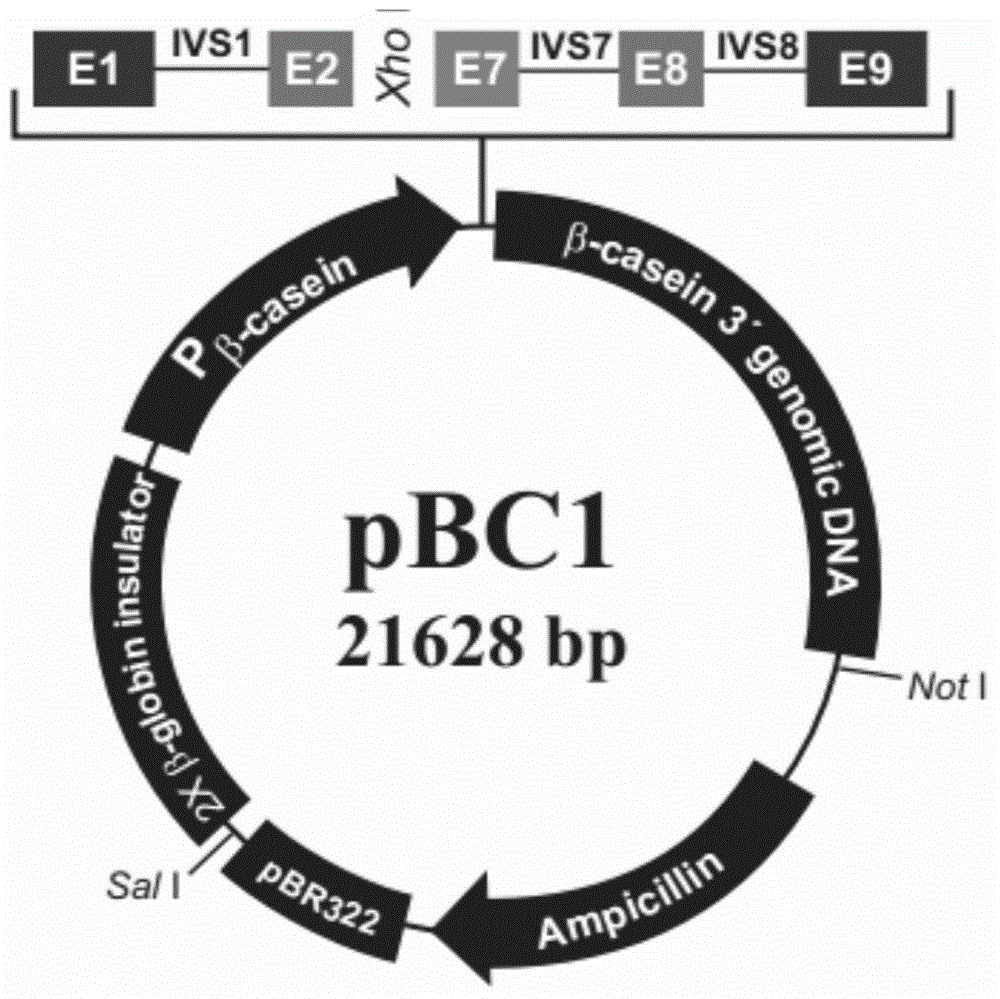
[0055] The pBC1 mammary gland-specific expression vector was purchased from Invitrogen. The structure of the vector is relatively simple, with only one ampicillin resistance gene and no multiple cloning site (MCS). There are only 3 available restriction sites, XhoI, NotI, and SalI, on the entire vector, and it is required that the target gene must be inserted into the XhoI site for normal expression ( figure 1 ). Therefore, only the single enzyme digestion method can be selected when constructing the vector.
[0056] 1. Cloning of the full-length HIOMT gene with XhoI restriction site
[0057] (1) Design of HIOMT full-length cloning primers
[0058] We designed upstream and downstream primers at the 5' and 3' non-coding regions of the HIOMT gene respectively, and introduced the recognition site CTCGAG of the XhoI restriction endonuclease at the 5' end of the upstream and downstream primer...
Embodiment 2
[0104] Example 2 Optimization of the method for constructing a single-enzyme-cut vector
[0105] In this example, 400 samples were subjected to PCR at different ligation times at 16°C, and the positive rates are shown in Table 1:
[0106] Table 1 The influence of different connection time on the positive rate
[0107] connection time 60min 90min 4h 16h positive rate 5.20% 8.33% 10.41% 3.12%
[0108] It can be seen from Table 1 that when the connection time is 4 hours, the positive rate reaches 10.41%, and when the connection time is 90 minutes, the positive rate reaches 8.33%. When the connection time is 90 minutes, the connection efficiency has been significantly improved. Taking time cost and connection efficiency into consideration, the present invention selects 90 minutes as the optimal connection time.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com