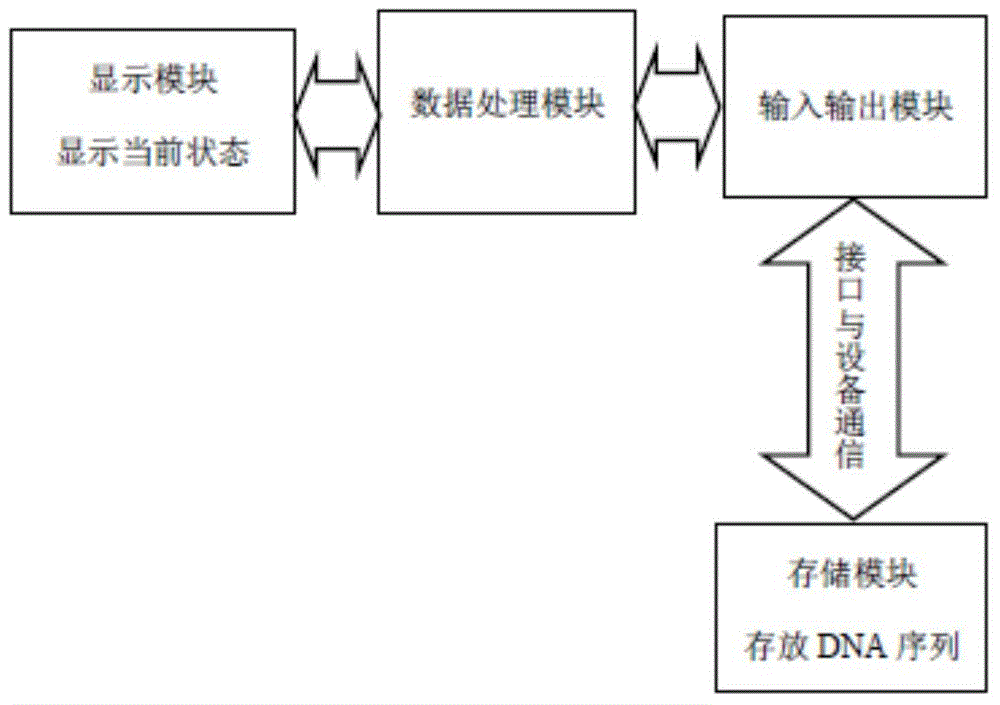
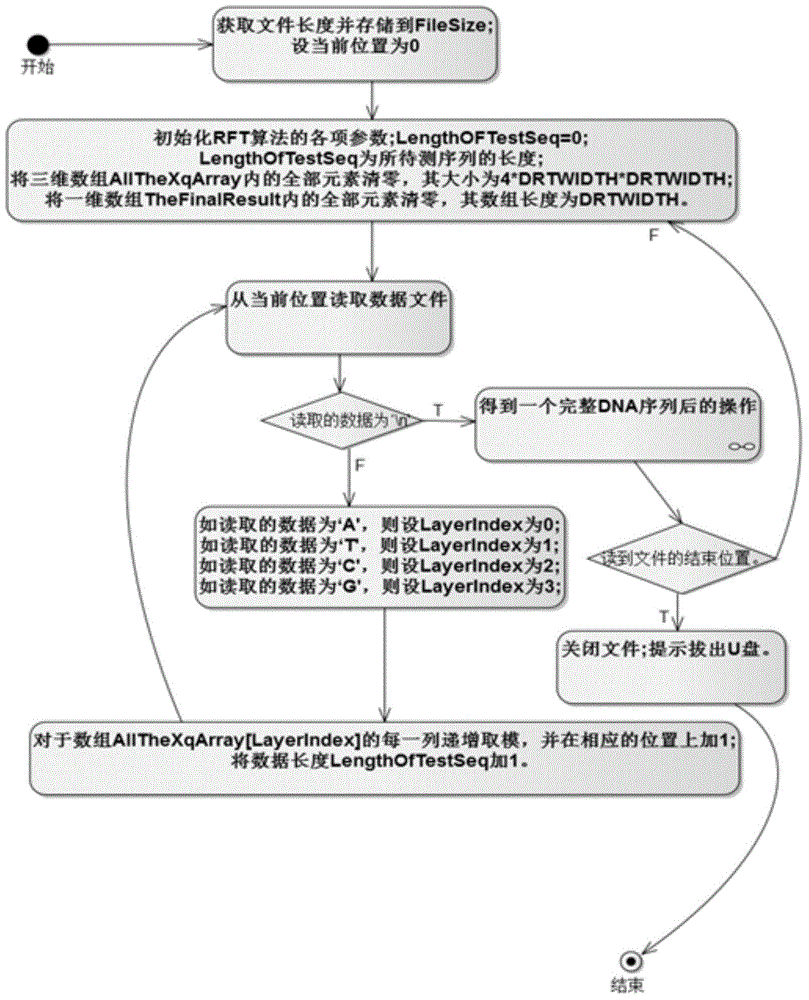
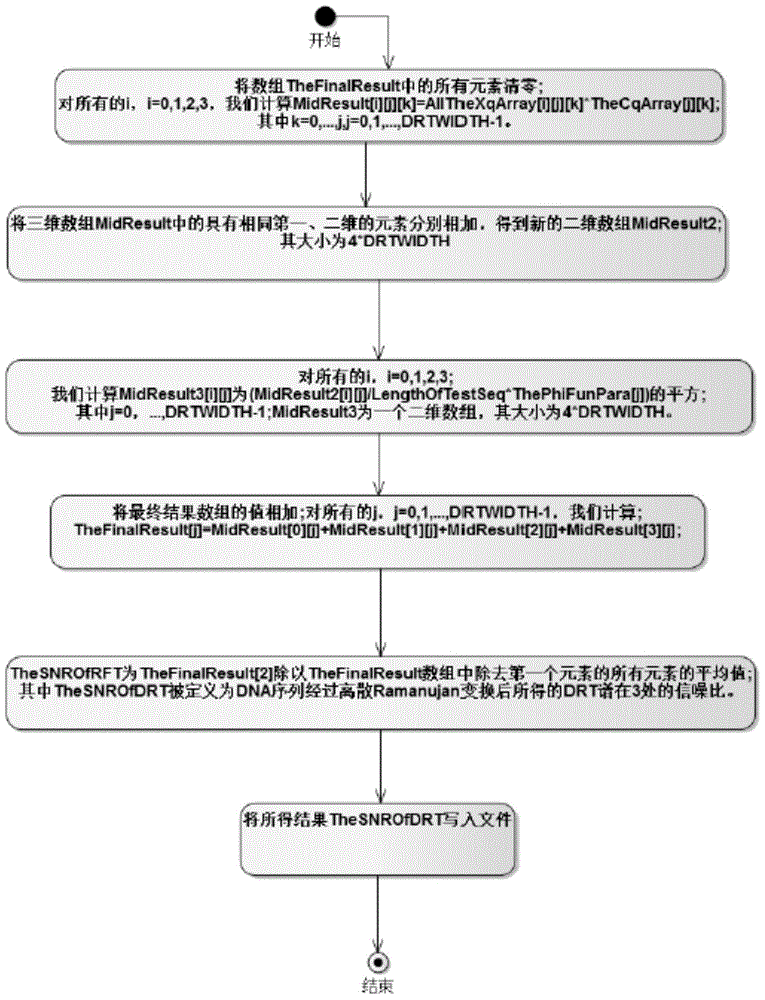
System for identifying coding region and non-coding region in DNA (Deoxyribose Nucleic Acid) gene sequence
A technology of DNA sequence and coding region, which is applied in the system field of identifying coding region and non-coding region in DNA gene sequence, which can solve the problems of floating-point calculation cost and excessive calculation time, and achieve the effect of small amount of calculation and high precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] Preferred embodiments of the present invention will be described in detail below in conjunction with the accompanying drawings.
[0038] Based on Ramanujan sum and Ramanujan coefficients, the embodiment proposes the concept of discrete Ramanujan transform spectrum. By using the Voss numerical representation, the character DNA sequence is mapped to the numerical DNA sequence, thereby obtaining the discrete Ramanujan spectrum of the numerical DNA sequence.
[0039] Since the spectrum of the discrete Fourier transform of protein-coding sequences has a significant 3-periodicity, this is the basis for the discrete Fourier transform to be used in DNA sequence analysis. It uses the signal-to-noise ratio of the sequence at N / 3 frequency as a measure, where N refers to the length of the sequence. The examples indicate that 3-periodicity can be identified by a prominent peak at 3 in the discrete Ramanujan spectrum of protein coding sequences. As a numerical measure, the signal-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com