A method for automatic correction of genome metabolic network model
A metabolic network and genome technology, which is applied in the field of automatic correction of genome metabolic network models, can solve problems such as the inability to provide reaction interval positioning, and achieve a comprehensive effect of correcting the model.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0046] The present invention will be further described below in conjunction with embodiment.
[0047] Taking the correction process of the rough model of the Spathaspora passalidarum NRRL Y-27907 genome as an example to illustrate, the loopholes in the rough model are automatically filled in, and the direction of the reaction and the interval of the reaction are determined. Specific steps are as follows:
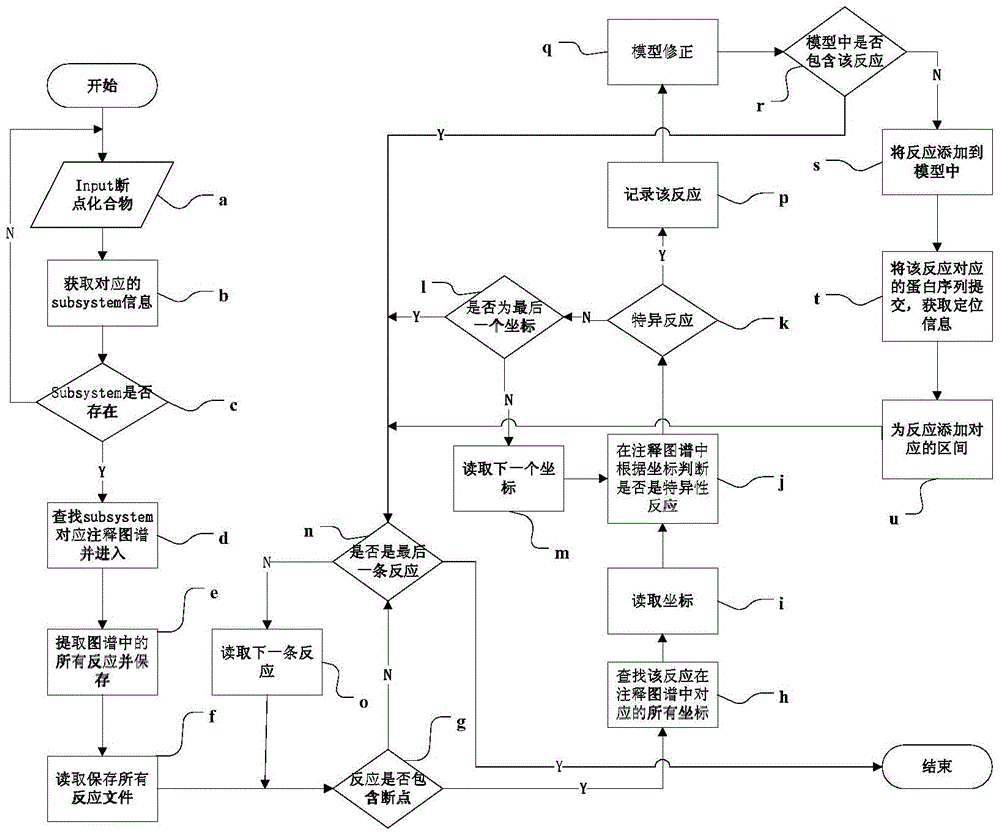
[0048] 1. According to the list of vulnerable metabolites returned by matlab, fill in the specific reaction of Spathaspora passalidarum NRRL Y-27907. figure 1 It is the flow chart of step 1, which shows the process of auto-filling breakpoints, and provides an intuitive description of the process of auto-filling breakpoints in the form of program flow, and can be accessed fromfigure 1 See the process of cyclically judging each compound and each reaction in turn.
[0049] (1A), import the rough model of genome metabolic network into the matlab software equipped with cobra too...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com