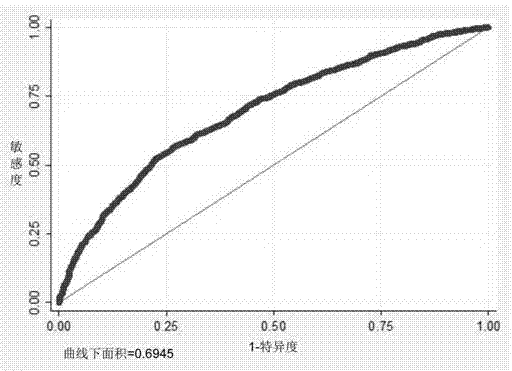
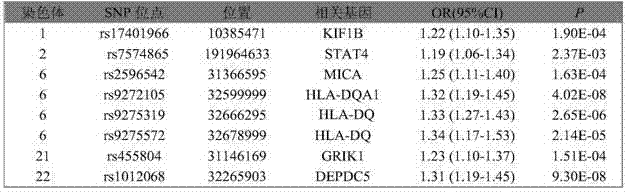
SNP marker related to primary hepatocellular carcinoma auxiliary diagnosis and application of SNP marker
A technology for auxiliary diagnosis and markers, applied in the field of SNP markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1 The collection of samples and the arrangement of sample data
[0065] The inventor collected a large number of blood samples from patients with liver cancer from Jiangsu and its surrounding areas from 2004 to 2013. After sorting out the sample data, the inventor selected 7323 samples that met the following criteria: whole genome chip scan and single SNP Experimental samples for Sequenom MassARRAY genotyping:
[0066] 1. Liver cancer patients with definite pathological diagnosis;
[0067] 2. Healthy controls matched with the age and sex of the cases;
[0068] The demographic data and clinical data of these samples were collected systematically.
Embodiment 2
[0069] Example 2 Whole Genome Scanning of SNPs in Peripheral Blood DNA
[0070] Among the above-mentioned 1538 eligible liver cancer patients and 1465 healthy controls, the two groups were matched in age and gender. The two groups of people were detected by the Human OmniExpress12v1 chip to obtain relevant results. The specific steps are:
[0071] 1. Add the hemolysis reagent to the leukocytes stored in the 2ml cryopreservation tube, mix it upside down and transfer it completely.
[0072] 2. Removal of red blood cells: Fill the 5ml centrifuge tube to 4ml with hemolysis reagent, mix by inverting, centrifuge at 4000rpm for 10 minutes, and discard the supernatant. Add 4ml of hemolysis reagent to the precipitate, invert and wash again, centrifuge at 4000rpm for 10 minutes, and discard the supernatant.
[0073] 3. Extract DNA: add 1ml extract solution to the precipitate (each 300ml contains 122.5ml 0.2M sodium chloride, 14.4ml 0.5M ethylenediaminetetraacetic acid, 15ml 10% sodiu...
Embodiment 3
[0082] Example 3 Sequenom MassARRAY genotyping of a single SNP
[0083] The SNPs found to be associated with the onset of liver cancer in the above genome-wide scan were detected in another 2112 liver cancer cases and 2208 healthy controls. The specific steps are as follows:
[0084] 1. Add hemolysis reagent (i.e. lysate, 40 parts) to the peripheral blood stored in the 2ml cryopreservation tube. Dilute the TrisHcl solution to 2000ml, the same below), turn it upside down and mix it completely.
[0085] 2. Removal of red blood cells: Fill the 5ml centrifuge tube to 4ml with hemolysis reagent, mix by inverting, centrifuge at 4000rpm for 10 minutes, and discard the supernatant. Add 4ml of hemolysis reagent to the precipitate, invert and wash again, centrifuge at 4000rpm for 10 minutes, and discard the supernatant.
[0086] 3. Extract DNA: Add 1ml extract solution to the precipitate (each 300ml contains 122.5ml 0.2M sodium chloride, 14.4ml 0.5M ethylenediaminetetraacetic acid, 15...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com