A kind of micro-RNA marker and its application
A marker and tiny technology, applied in the field of biomedical testing, can solve problems such as poor diagnosis and prognosis of CTEPH, and achieve good tissue specificity and improve sensitivity and specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Primer Design
[0026] According to the sequence of hsa-miR-20a-5p, primers and probe sequences were designed. The specific sequence is as follows:
[0027] The reverse transcription primer sequence SEQ ID No.2 of hsa-miR-20a-5p:
[0028] GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT;
[0029] The forward primer sequence SEQ ID No.3 of hsa-miR-20a-5p:
[0030] GCGCGCTAAAGTGCTTATAGTGC;
[0031] The reverse primer sequence SEQ ID No.4 of hsa-miR-20a-5p:
[0032] GTGCAGGGTCCGAGGT;
[0033] The forward primer sequence of U6 is SEQ ID No.5: CTCGCTTCGGCAGCACA;
[0034] The reverse primer sequence of U6 is SEQ ID No.6: AACGCTTCACGAATTTGCGT;
[0035] The above primers were synthesized by GenScript Biotechnology Co., Ltd.
Embodiment 2
[0036] Example 2 total RNA extraction
[0037] (1) Take the blood sample (which has been added with Biotech lysate) and let it stand on ice for 5 minutes.
[0038] (2) Add 200 μl of chloroform, vortex and mix well, then place at room temperature for 2-3 minutes. Centrifuge at 12,000 g for 15 min at 4°C.
[0039] (3) Absorb the upper aqueous phase into another centrifuge tube. NOTE: Never pipette the middle interface.
[0040] (4) Add an equal amount of isopropanol, mix well, and precipitate at -20°C.
[0041] (5) Centrifuge at 12,000g at 4°C for 10 minutes, discard the supernatant, and sink the RNA to the bottom of the tube.
[0042] (6) Add 1ml of 75% ethanol to suspend the precipitate.
[0043] (7) Centrifuge at 12,000 g for 5 min at 4°C, and discard the supernatant as much as possible.
[0044] (8) Air dry at room temperature or vacuum dry for 5-10min. Note: RNA samples should not be too dry, otherwise it will be difficult to dissolve.
[0045] (9) with 20μl DEPC H ...
Embodiment 3
[0047] Example 3 Prediction of miRNAs associated with CTEPH
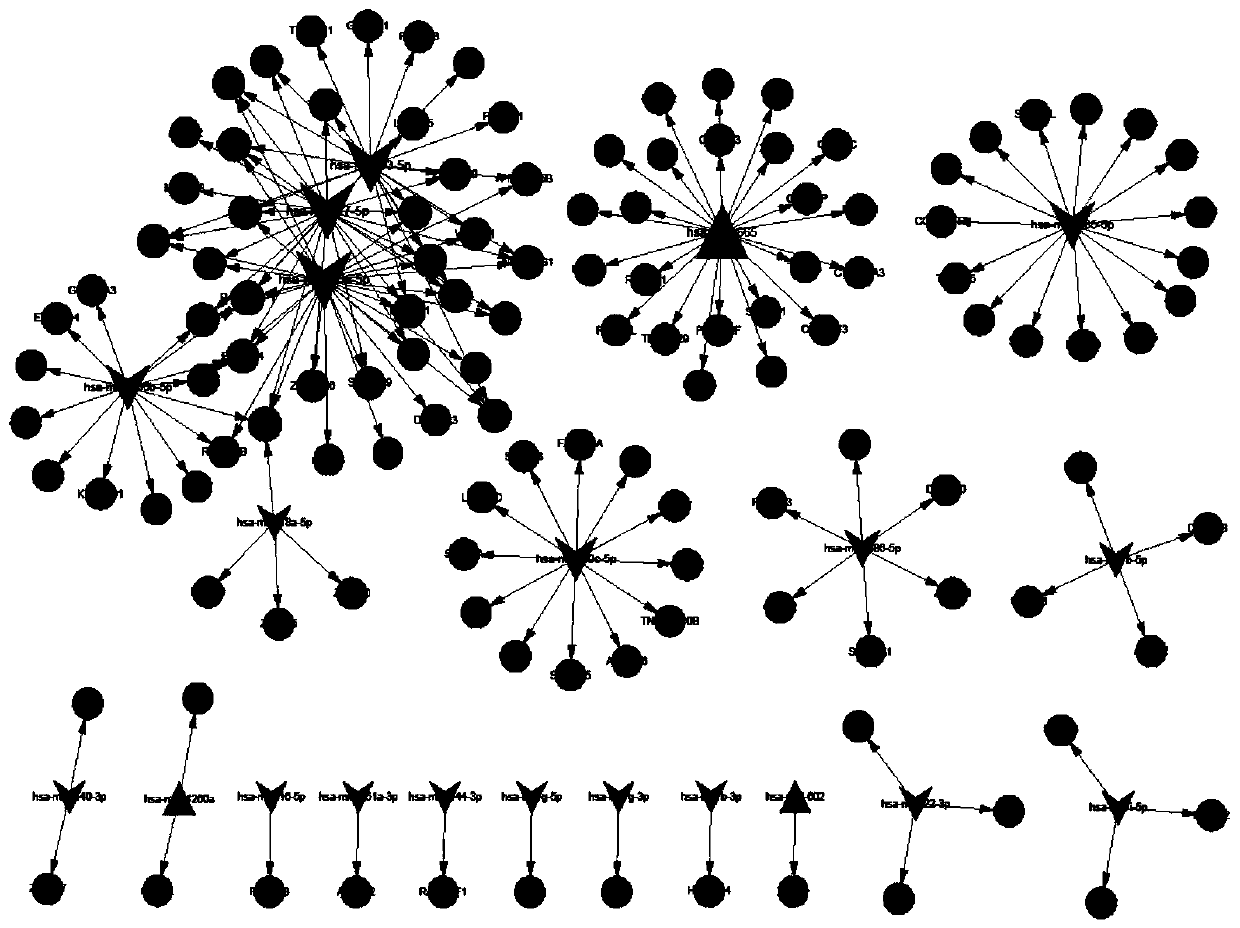
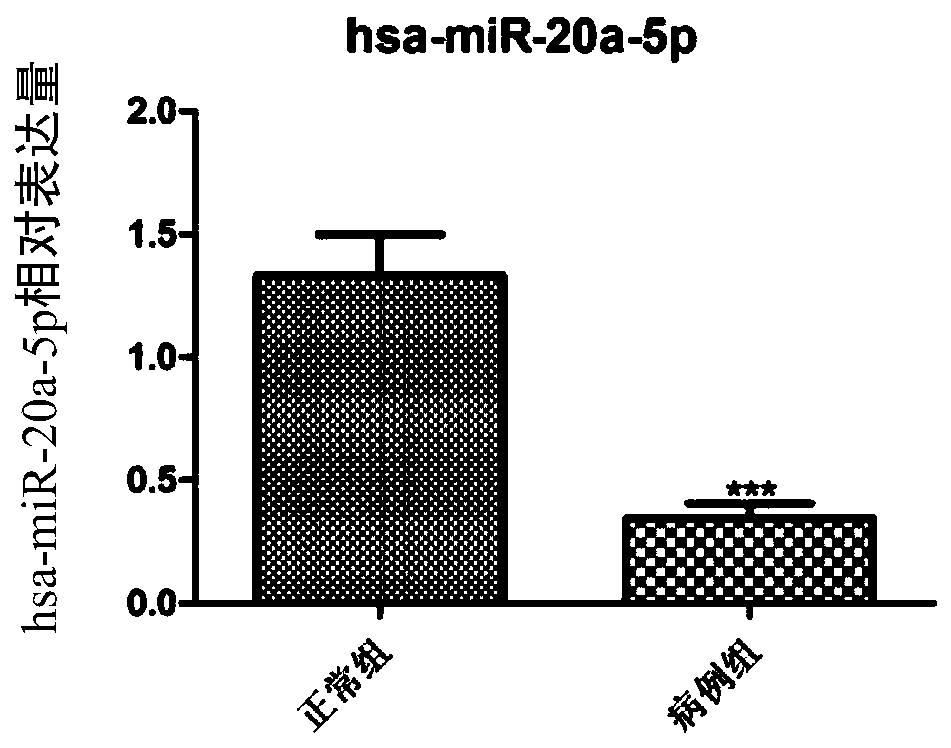
[0048] Using bioinformatics analysis technology, 169 miRNA-target relationship pairs were predicted by miRWalk2.0 software, including 21 miRNAs, 3 up-regulated miRNAs, and 18 down-regulated miRNAs; the above predicted miRNA-target relationship pairs were used to construct a regulatory network Figure, miRNA-target regulatory network see figure 1 , a total of 147 nodes, see the specific results figure 1 . Among them, hsa-miR-20a-5p is a down-regulated miRNA associated with CTEPH disease.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com