Application of miRNA-26a
A miRNA-26a, marker technology, applied in the field of use, can solve the problem of undiscovered miRNA functions, and achieve the effects of improving sensitivity and specificity, easy detection, and accurate quantification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Screening of miRNAs associated with human liver cancer
[0052] 1.1 Collection of samples and collation of sample data
[0053] The inventor collected a large number of peripheral blood samples from patients with liver cancer and benign liver diseases in Beijing Cancer Hospital in January 2013 (the samples used for research were collected at the same time, sampled, subpackaged, and stored under uniform conditions). The inventor selected 20 cases of plasma from patients with liver cancer (hepatitis B-cirrhosis-liver cancer) randomly collected in the same hospital and as far as possible in the same department, and 20 cases of plasma from patients with benign liver diseases randomly collected in the same hospital and the same period as the experimental group. Try to avoid taking plasma samples from people with a family history of liver cancer for miRNA chip detection.
[0054] 1.2 miRNA microarray detection
[0055] 1.2.1 Isolation and purification of exosome ...
Embodiment 2
[0072] Example 2 QPCR verification of differentially expressed miRNA-26a
[0073] According to the detection results of the miRNA chip, miRNA-26a was selected for large sample QPCR verification. According to the method of sample collection and sample data arrangement in Example 1, 100 samples were selected from the liver cancer group and the benign liver disease group.
[0074] 2.1 The RNA extraction process is the same as in Example 1.
[0075] 2.2 Reverse transcription: mix 10pg-1μg total RNA template with 2μl 10* buffer, 2μl dATP (10mM), 0.5μl polyA polymerase, 0.5μl ribonuclease (RNase) inhibitor and ribonuclease free water (RNase free water ) were mixed, the final volume was 20 μl, and incubated at 37° C. for 1 h. Then add 1 μl 0.5 μg / μl Oligo(dT) specific RT primer to the reaction tube, incubate at 70°C for 5 minutes and immediately incubate on ice for at least 2 minutes to break the secondary structure of RNA and primers. Finally, mix the above 20 μl reaction mixture...
Embodiment 3
[0077] Example 3 Analyzing the diagnosis of miRNA on the pathogenesis of liver cancer
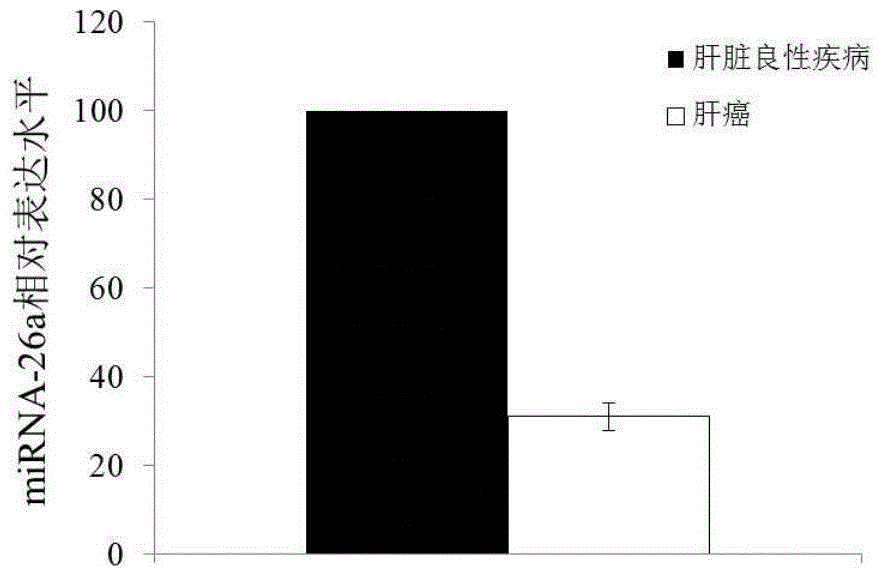
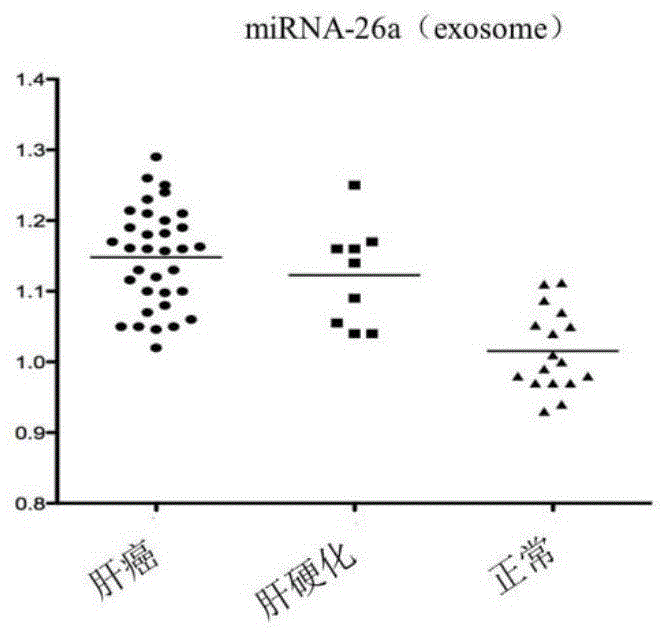
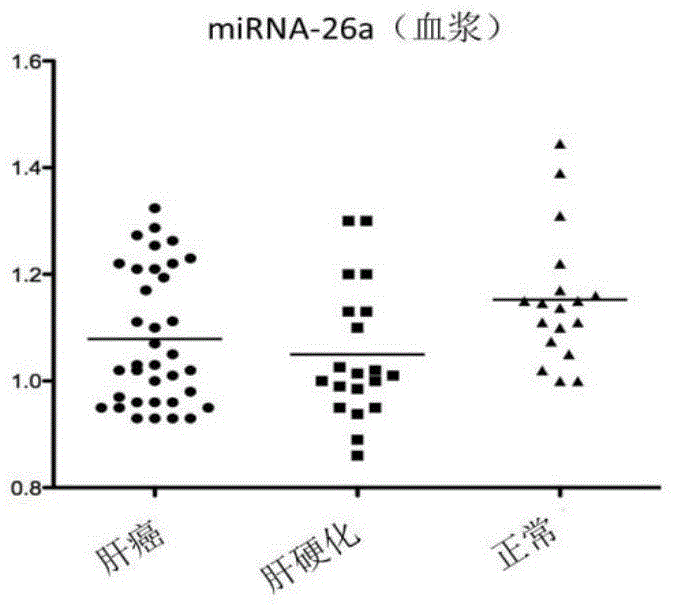
[0078]The exosome-derived miRNA-26a was used as the research object, and compared with plasma free miRNA in the sensitivity and specificity of the control group and liver cancer group. Taking liver cancer patients as the experimental group, liver cirrhosis patients and normal people as the control group, exoquic reagent was used to extract the exosome in the plasma (the steps are the same as in Example 1). 40 cases of liver cancer plasma samples, 20 cases of liver cirrhosis patients and 20 cases of normal human plasma samples were selected for miRNA-26a QPCR experiment, the CT value of each sample was obtained, and the scatter plot was made by using the ratio of sample miRNA-26a CT value to U6CT value (Figure 2) The experiment proved that the exosome group had better aggregation than the plasma group. The exosome group experimental results had the same expression trend of miRNA-26a in liver...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com