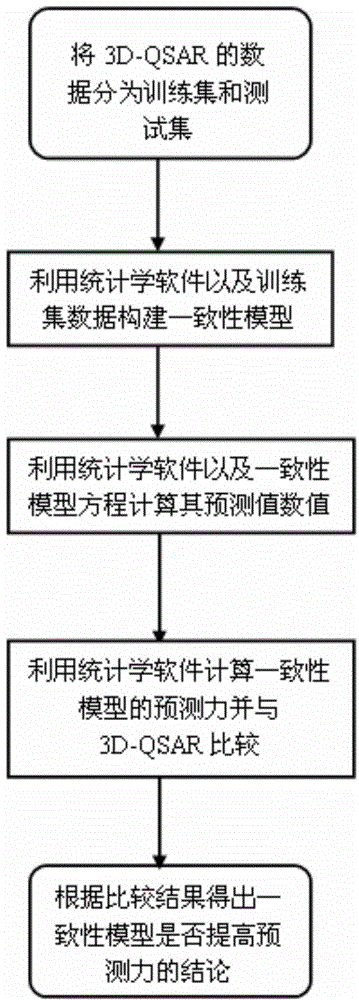
Consistency model building method based on 3-dimensional quantitative structure-activity relationship model
A technology of structure-activity relationship and construction method, applied in the field of bioinformatics, can solve problems such as not being able to contain all the information of compounds, and the prediction accuracy of a single model is not high, and achieve the effect of improving the prediction ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The present invention relates to the use of HIV-1 integrase inhibitor compound molecules derived from (POLANSKI J, ZOUHIRI F, JEANSON L, DESMAELE D, D'ANGELO J, MOUSCADET J F, GIELECIAK R, GASTEIGER J, LE BRET M.Use of the Kohonen neural network for rapid screening of ex vivo anti-HIV activity of styrylquinolines[J].Journal of Medicinal Chemistry,2002,45(21):4647-4654; NORMAND-BAYLE M,BENARD C,ZOUHIRI F,MOUSCADET J F,LEH H ,THOMAS C M,MBEMBA G,DESMAELE D,D'ANGELO J.New HIV-1replication inhibitors of the styryquinoline class bearing aroyl / acyl groups at the C-7position:Synthesis and biological activity[J].Bioorganic&Medicinal Chemistry Letters,2005,15 (18):4019-4022; LEONARD J T,ROY K.Exploring molecular shape analysis of styrylquinoline derivatives as HIV-1 integrase inhibitors[J].Eur J Med Chem,2008,43(1):81-92;MOUSCADET J F,DESMAELE D.Chemistry and Structure-Activity Relationship of the Styrylquinoline-Type HIV Integrase Inhibitors[J].Molecules,2010,15(5):3048-3078; O...
Embodiment 2
[0080] According to the data of HIV-1 entry inhibitor 3D-QSAR model in the literature (C.Teixeira, N.Serradji, F.Maurel, F.Barbault, Docking and 3D-QSAR studies of BMS-806analogs as HIV-1gp120entry inhibitors, European Journal of Medicinal Chemistry 44(2009) 3524–3532.), Constructing Consistency Models.
[0081] Using biostatistical methods, the 3D-QSAR model training set compound molecular pIC 50 The predicted value of is used as the independent variable, and the experimental measurement value is used as the dependent variable. The formula for obtaining the consistency model is shown in formula 12.
[0082] pIC 50 =0.736×P CoMFA +0.582×P CoMSIA -0.111 (12)
[0083] Using statistical software, substitute the pIC50 value of CoMFA and CoMSIA prediction set compound molecules into formula (12), and calculate the predicted value of the consistency model prediction set molecules.
[0084] The predicted value of the consistency model test set is used as the independent variable...
Embodiment 3
[0086]In this example, the data of 3D-QSAR models of 90 HIV inhibitors in recent years were collected, and the predicted values and experimental values of the 3D-QSAR models in the training set and prediction set in the literature were used to construct the consistency model. Since the value of Topomer CoMFA has a strong correlation with the values of CoMFA and CoMSIA models, after the consistency model is constructed, only the consistency model is compared with the values of CoMFA and CoMSIA models, and the comparison is based on the cross-validation correlation of the original model. Coefficient q 2 The size is divided into 0.52 ≤0.6; 0.62 ≤0.7; 0.72 ≤0.8; 2 >0.8 four categories. After eliminating suspicious values, analyze them respectively according to the above four categories and the five methods without classification.
[0087] The results of testing the consistency model and the CoMFA model with the paired t test method are shown in Table 4. The cross-valida...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com