ISSR(Inter-simple Sequence Repeat)-PCR(Polymerase Chain Reaction) molecular marker method for macromitrium gymnostomum
A technology of Moss de dentatus and molecular marker, applied in the field of molecular biology, can solve the problem of not many bryophytes research, and achieve the effects of great scientific value and application value, high stability and high definition
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Genomic DNA was extracted from Moss dentata.
Embodiment 2
[0028] Screening and optimization of the amplification system
[0029] 1. Orthogonal experimental design of exploratory ISSR-PCR reaction system
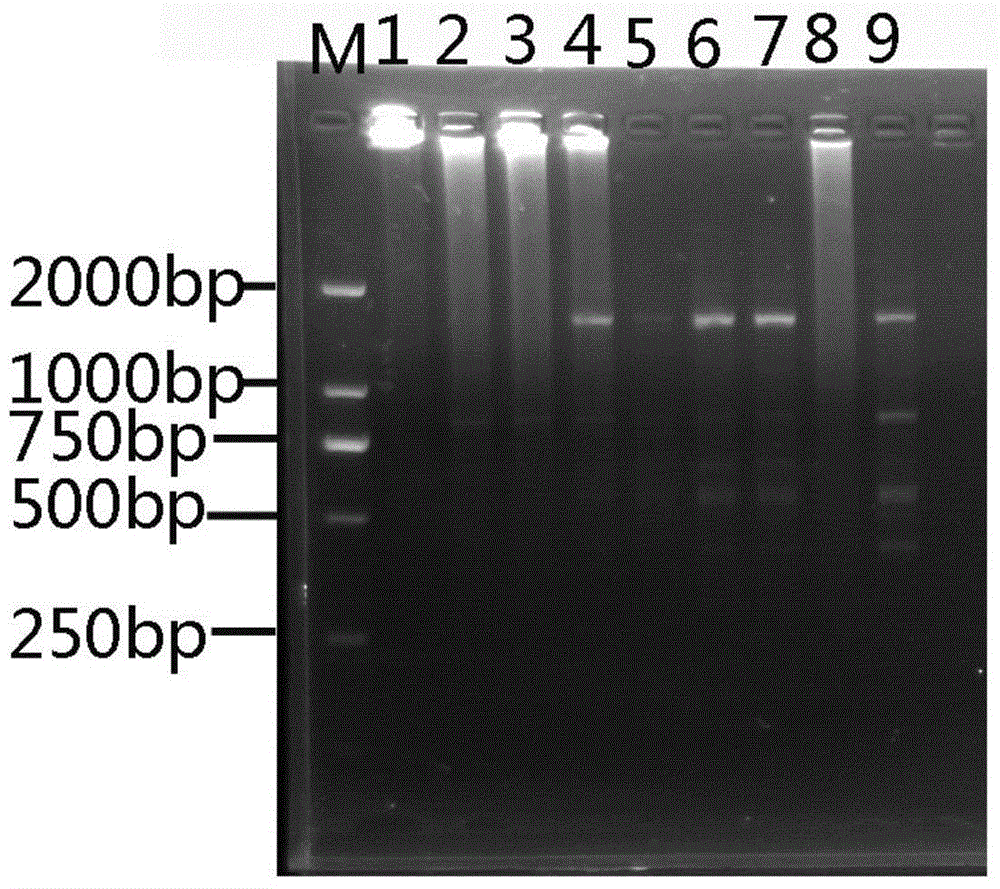
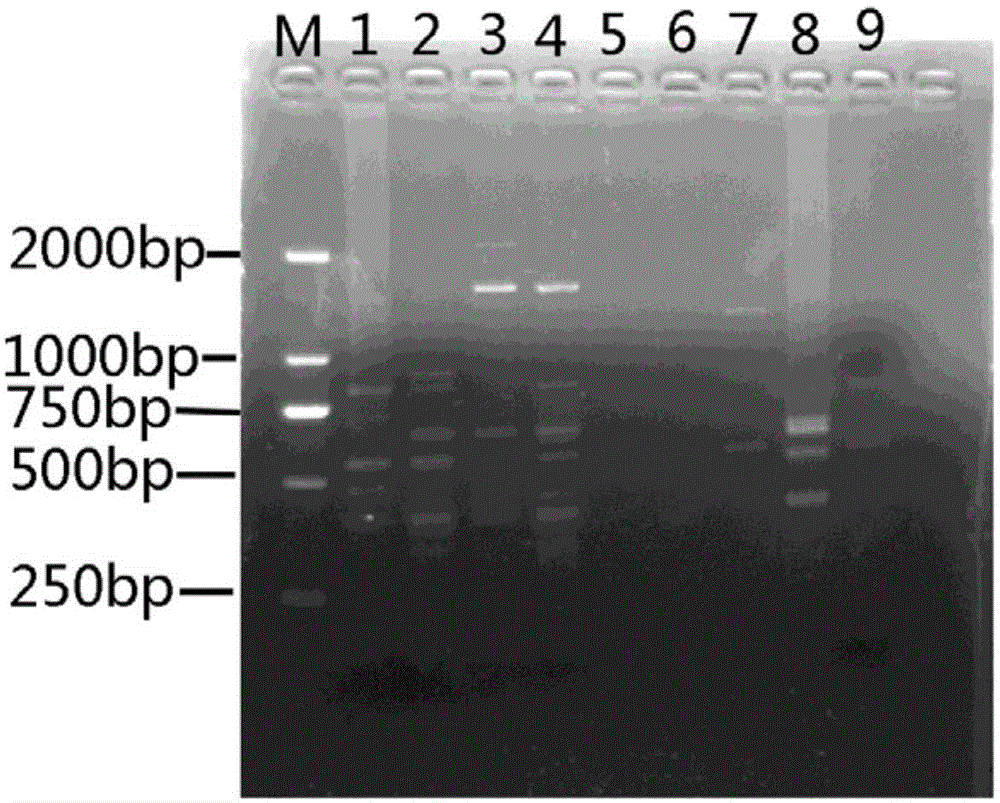
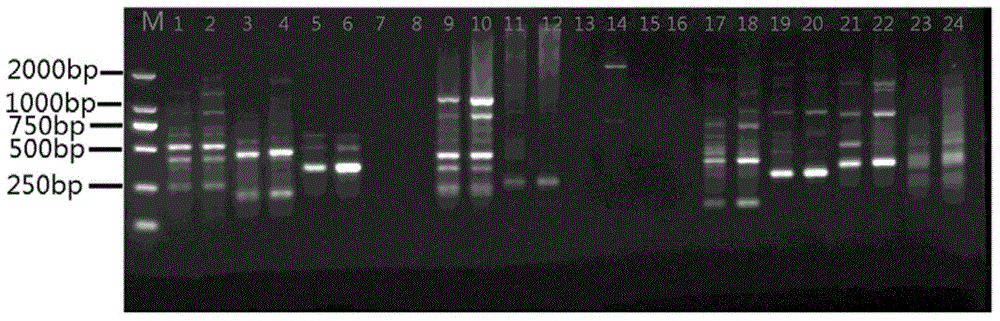
[0030] Orthogonal optimization design screens the optimal response system of ISSR (four factors and three levels), choose L 9 (3 4 ) orthogonal table, design the factor-horizontal orthogonal design experiment table of each component of the PCR amplification system (see Table 2 and Table 3 for the scheme). Using UBC834 as a primer, the 9 systems in Table 3 were used to perform ISSR amplification on the genomic DNA of Moss dentata. The reaction system is 30 μL. Except for the factors listed in the table, each tube contains 3 μL of 10×LA PCR buffer and 50 ng of DNA template. The amplification program was: annealing at 94°C for 4min, followed by 40 cycles of 94°C for 45S, 53°C for 45S, and 72°C for 1min, and finally extension at 72°C for 10min, and storage at 10°C.
[0031] Table 1 Primers and sequences for ISSR amplification
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com