Primer for simultaneously detecting XRCC1, ERCC, GSTP1 and GSTM1 gene polymorphism, and method thereof
A technology of gene polymorphism, -TCTGACTCCCCTCCAGATT-3, applied in biochemical equipment and methods, microbe measurement/inspection, etc., can solve the problems of high sensitivity and lack of specificity, and achieve high sensitivity, good accuracy, and improved The effect of detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Primer
[0024]The inventor designed a large number of primers for the gene polymorphism sites of XRCC1 / ERCC1 / ERCC2 / GSTP1 / GSTM1, and through the optimization and comparison of the primer reaction conditions, the PCR reaction conditions with good specificity, no cross-reaction and no cross-reaction were selected. very close primer. The primers provided by the present invention include PCR amplification primers and SNaPshotPCR primers, and the PCR amplification primers and SNaPshotPCR primers are corresponding. All primer sequences provided by the present invention are compared through UCSC database, and there is no known SNP site.
[0025]
Embodiment 2
[0026] The specificity of embodiment two primers
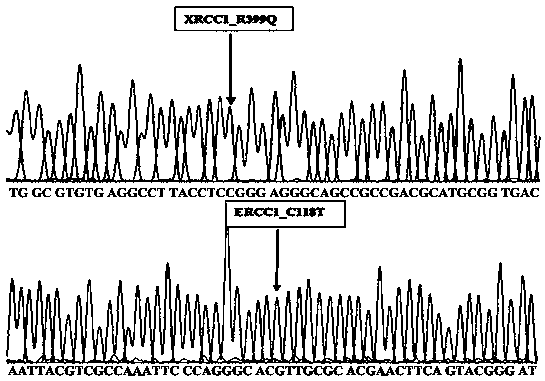
[0027] The primers provided by the present invention were blasted in UCSC. The amplified fragment of the XRCC1_R399Q primer was located at chr19:44055651-44055888, with a length of 238bp; chr19:45854837+45855007, the length is 171bp; the GSTP1_I105V primer amplified fragment is located at chr11:67352602-67352777, the length is 176bp; the GSTM1_Loss primer amplified fragment is located at chr1:110232920-110233138, the length is 219bp; the results are as follows Figure 1 to Figure 2 As shown, the amplified fragments of all primers covered the corresponding detection sites, and there were no other homologous genes.
[0028] The PCR amplification primers in Table 1 were used to amplify and Sanger sequence the test samples respectively. The sequencing results showed that the fragments amplified by each primer were consistent with the reference sequence of the XRCC1 / ERCC1 / ERCC2 / GSTP1 / GSTM1 gene. The results were as follows Figu...
Embodiment 3
[0036] Example 3 Specificity of the method for detecting XRCC1 / ERCC1 / ERCC2 / GSTP1 / GSTM1 gene polymorphism
[0037] The specificity of this assay is defined as the negative coincidence rate. In this test, the detection of XRCC1_R399QG / G genotype is defined as a negative result, the detection of ERCC1_C118TC / C genotype is defined as a negative result, the detection of ERCC2_L751QA / A genotype is defined as a negative result, and the detection of GSTP1_I105VA / A genotype is defined as a negative result. It was defined as a negative result, and the detection of GSTM1 non-deleted genotype was defined as a negative result.
[0038] The SNaPshot method provided by the present invention was used to detect 9 samples, and at the same time, the ARMS method or Sanger sequencing method was used for verification. The detection results of the SNaPshot method are consistent with those of the ARMS method or the Sanger sequencing method, as shown in Table 4. The specificity of this detection m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com