Specific primer and parting method for II-type MHC gene for anti-bacterial potential detection on crested ibis
A typing method and genotyping technology, which can be applied to biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0043] 1. Sample preparation: Crested ibis blood and tissue samples came from artificial populations such as Deqing, Sado, Beijing, Henan, Yangxian and Louguantai.
[0044]2. DNA extraction: phenol chloroform method to extract blood and tissue DNA.
[0045] 3. Primer synthesis: According to the obtained crested ibis class II-MHC gene sequence, specific primers for the II-A1 site were designed to amplify exon 2 for genotyping of the modified site, and BGI synthesized primers. The primer sequences are as follows:
[0046]
[0047] 4. Primer amplification: apply the above amplification primers to PCR respectively to amplify the extracted sample DNA. Set the PCR thermal cycle annealing temperature range to 64°C.
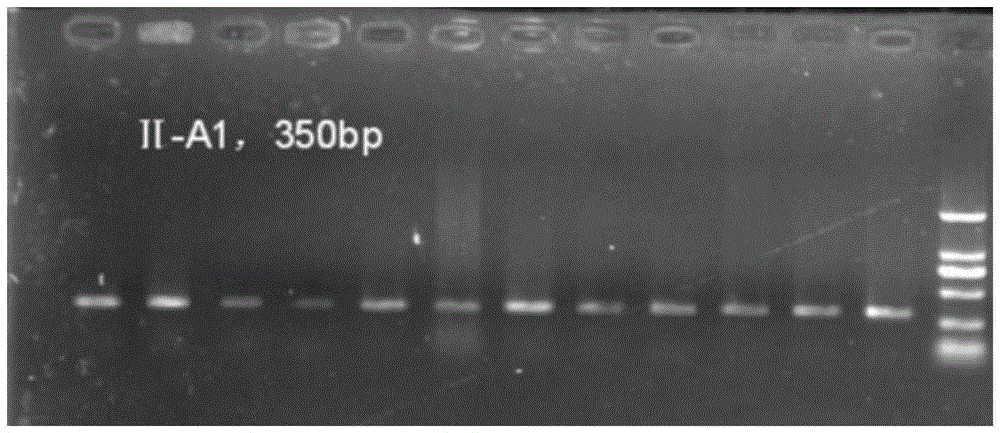
[0048] 5. Amplification result detection: take 2 ul of the above-mentioned PCR amplification products and carry out agarose gel electrophoresis, the gel concentration is 1.5%. The result is as figure 1 As shown, this primer can effectively amplify the target band,...
example 2
[0062] 1. Sample preparation: Crested ibis blood and tissue samples came from artificial populations such as Deqing, Sado, Beijing, Henan, Yangxian and Louguantai.
[0063] 2. DNA extraction: phenol chloroform method to extract blood and tissue DNA.
[0064] 3. Primer synthesis: According to the obtained crested ibis class II MHC gene sequence, specific primers for the II-B1 site were designed to amplify exon 2, and the primers were synthesized by BGI. The primer sequences are as follows:
[0065]
[0066] 4. Primer amplification: apply the above amplification primers to PCR respectively to amplify the extracted sample DNA. Set the PCR thermal cycle annealing temperature range to 63°C.
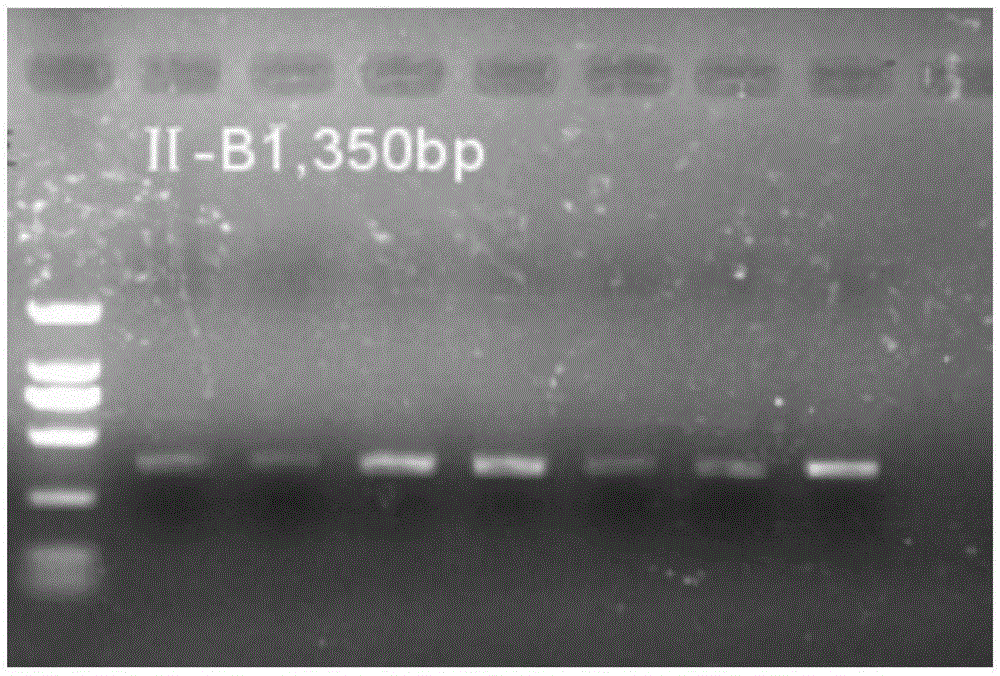
[0067] 5. Amplification result detection: take 2 ul of the above-mentioned PCR amplification products and carry out agarose gel electrophoresis, the gel concentration is 1.5%. The result is as figure 2 As shown, this primer can effectively amplify the target band, the fragment size of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com