Methylation marker for early detection of cancer and detection method thereof
A methylation marker, early detection technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. The effect of improving quality of life, sensitivity and specificity, and prolonging survival
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
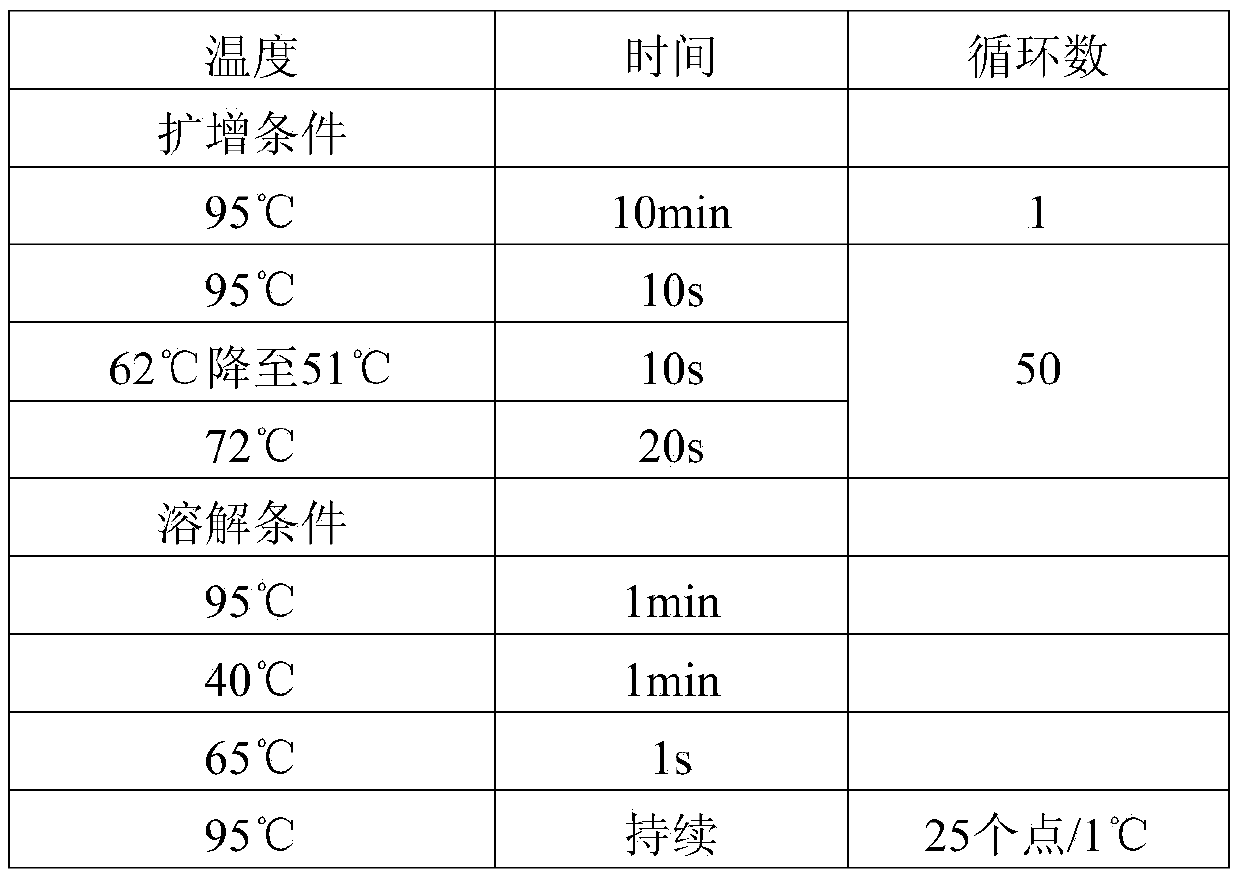
[0032] A set of specific primers designed for methylation markers used in the diagnosis of lung cancer:
[0033] According to the whole human genome sequence released by NCBI (National Center for Biotechnology Information), it was designed using PrimerPremier3.0 and MethylPrimerExpressv1.0, and synthesized by Shanghai Sangong Co., Ltd. The upstream primer and downstream primer of the PCDHGB6 gene are respectively shown in SEQIDNO.1 and SEQIDNO.2; the upstream primer and downstream primer of the HOXA9 gene are respectively shown in SEQIDNO.3 and SEQIDNO.4; the MGMT The upstream primer and downstream primer of the gene are respectively shown in SEQIDNO.5 and SEQIDNO.6; the upstream primer and downstream primer of the MicroRNA-126 gene are respectively shown in SEQIDNO.7 and SEQIDNO.8.
Embodiment 2
[0035] Application of the kit of the present invention in detecting the methylation level of human lung cancer:
[0036] The Master used in the present invention was purchased from Roche Co., Ltd.; the DNA extraction kit was QIAmpDNAMiniKit (QIAGEN); the transformation kit was EZDNAMethylation kit (ZymoResearch, Orange, CA, USA); other reagents were domestic analytical reagents.
[0037] The biological materials used in the present invention are all from Shanghai Zhongshan Hospital.
[0038] method:
[0039] 1. Biological samples:
[0040] Tumor tissues and paired normal tissues of 54 lung cancer patients.
[0041] 2. Extraction and transformation of tissue DNA:
[0042]Take about 25mg of lung cancer tissue or normal tissue, cut it into pieces with surgical scissors after autoclaving and put it into a 1.5ml centrifuge tube, use the DNA extraction kit to extract the DNA of the sample to be tested, and then use the transformation kit to transform the extracted DNA .
[0043...
Embodiment 3
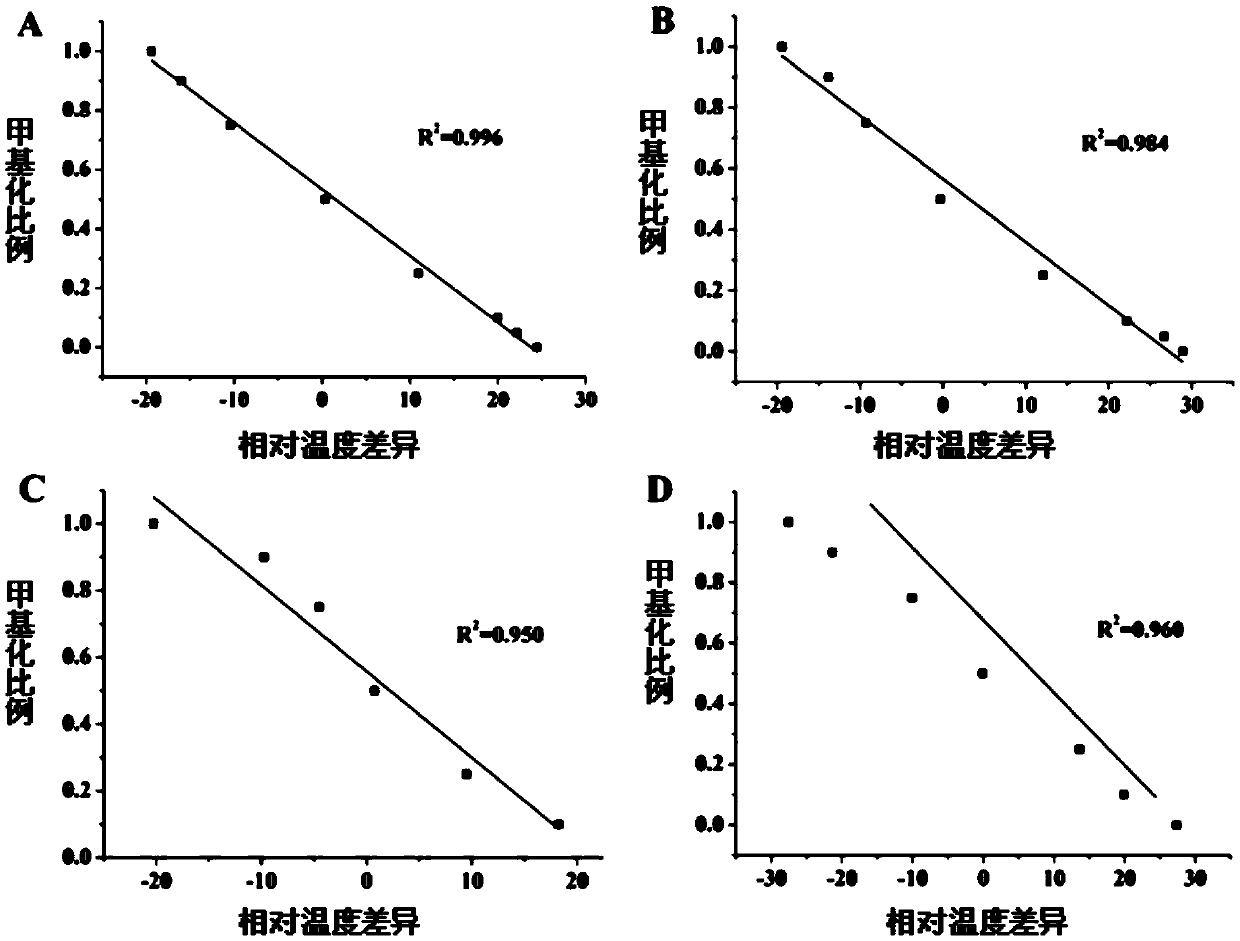
[0055] Operating characteristic curve analysis (ROC curve analysis):
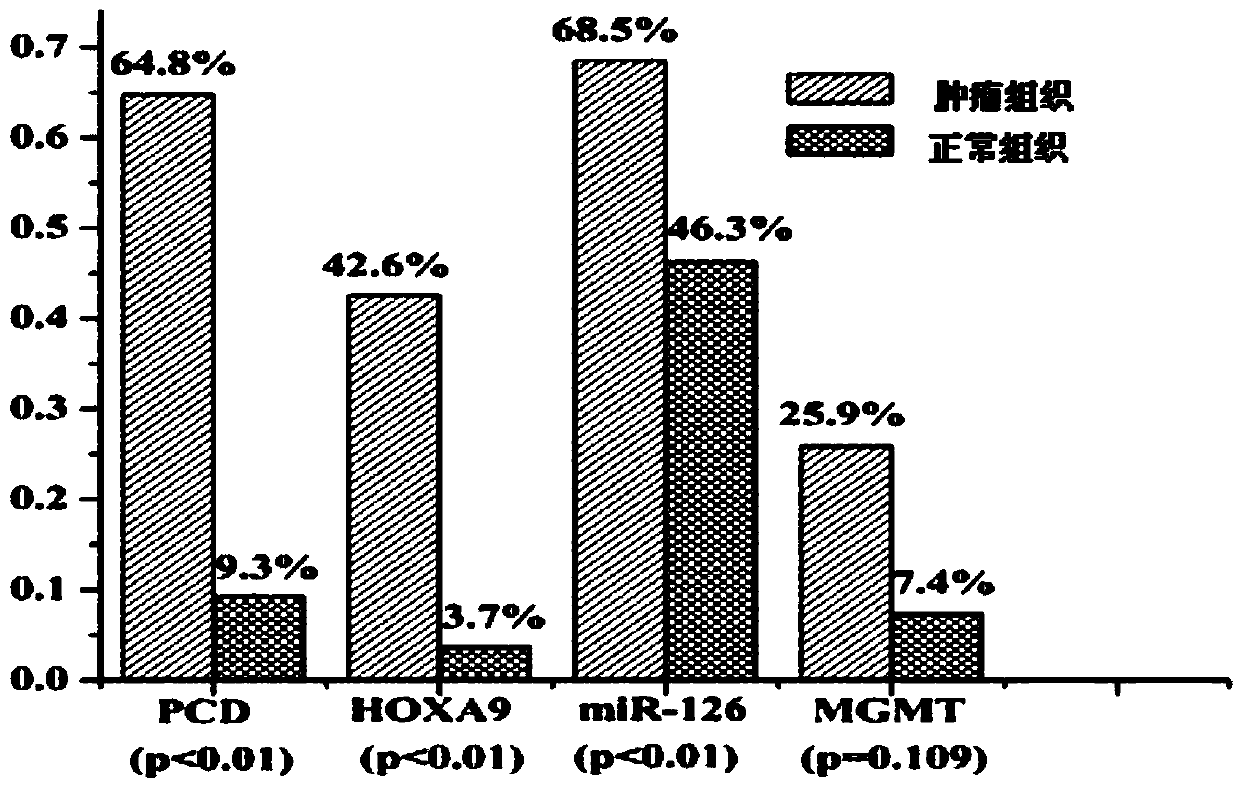
[0056] A ROC curve was constructed to compare the diagnostic ability of the four methylation markers in distinguishing tumor tissue from normal tissue in patients with lung cancer. The areas under the ROC curve of the four markers were as follows: PCDHGB6, 0.796; HOXA9, 0.694; MGMT, 0.594; Mir-126, 0.658. Under the optimal cutoff value, the sensitivity and specificity of the genes are as follows: PCDHGB6, 66.7% and 90.7%; HOXA9, 42.6% and 96.3%; MGMT, 25.9% and 94.4%; Mir-126, 38.9% and 90.7% . The combined detection AUC of these four markers reached 0.891, and the sensitivity and specificity were 85.2% and 81.5% respectively (see Table 3). These results indicated that the combined detection of lung cancer by the four methylation markers has relatively high sensitivity and specificity.
[0057] Table 3 ROC curve analysis of methylation markers
[0058]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com