A method for realizing scarless homologous recombination of Escherichia coli genes
A technology of Escherichia coli and homologous recombination, applied in recombinant DNA technology, biochemical equipment and methods, viruses/phages, etc., can solve problems such as the negative impact of accuracy and recombination efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
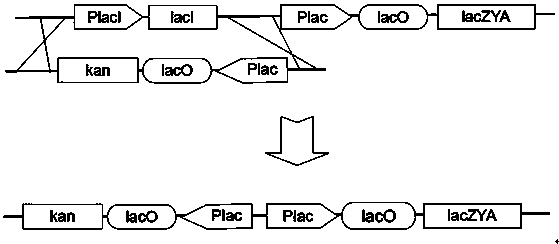
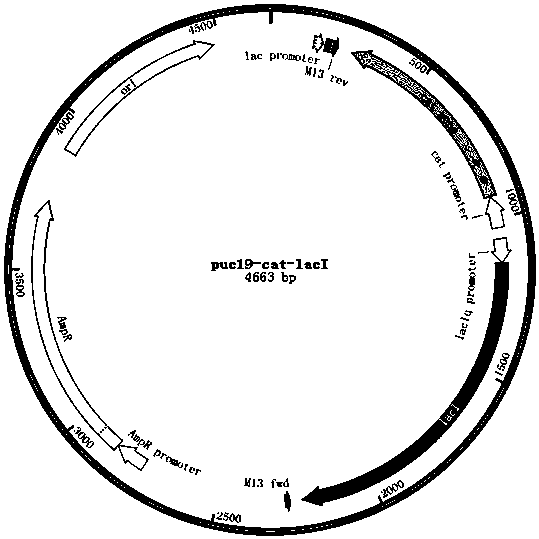
[0022] Example 1 Construct the kan operon of the kanamycin resistance gene initiated by the lac operator sequence, and replace the wild-type lacI gene and its promoter of Escherichia coli DH5α by homologous recombination. The recombinant plasmid pUC19-cat-PlacIq-lacI composed of a traceless recombination tool gene string composed of the lacI repressor gene in reverse, uses the cat-PlacIq-lacI tool gene string to replace the natural promoter of the talB gene in E. coli DH5α with trc promoter, the steps are as follows:
[0023] 1.1. The plasmid pKD46 was electrotransformed into Escherichia coli DH5α, and the strain that successfully introduced the plasmid was obtained by screening with ampicillin plate.
[0024] 1.2. Use the upstream and downstream primers lacI-Plac-kan-F and lacI-kan-R (DNA sequences shown in SEQ ID NO.1 and SEQ ID NO.2 respectively) to obtain Plac-lacO- For the kan operon gene, its homologous recombination was used to replace the wild-type lacI gene and its p...
Embodiment 2
[0030] Example 2 Construct the kan operon of the kanamycin resistance gene initiated by the lac operator sequence, and replace the wild-type lacI gene and its promoter of Escherichia coli BL21 (DE3) by homologous recombination, and construct a The recombinant plasmid pUC19-cat-PlacIq-lacI, which is composed of a traceless recombination tool gene string composed of cat and lacI repressor gene in reverse, uses the cat-PlacIq-lacI tool gene string to integrate the talB gene in Escherichia coli BL21 (DE3) Replace the natural promoter with the trc promoter, the steps are as follows:
[0031] 2.1. The plasmid pKD46 was electrotransformed into Escherichia coli BL21 (DE3), and the strains successfully introduced with the plasmid were obtained by screening with ampicillin plate.
[0032] 2.2. Use the upstream and downstream primers lacI-Plac-kan-F and lacI-kan-R (DNA sequences shown in SEQ ID NO.1 and SEQ ID NO.2 respectively) to obtain Plac-lacO- from plasmid pKD4 by pcr amplification...
Embodiment 3
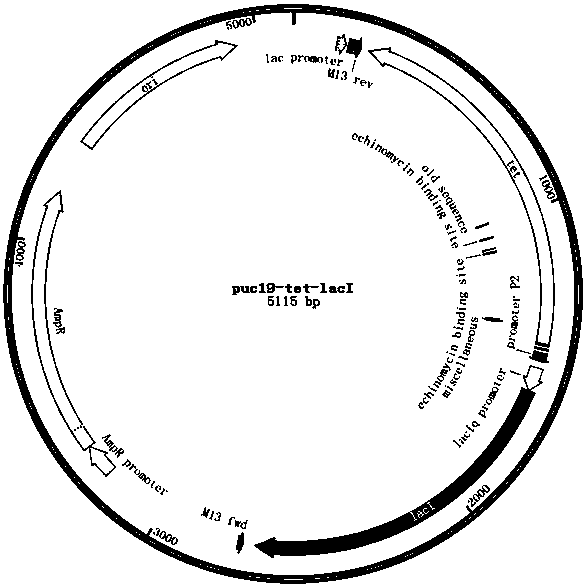
[0038] Example 3 Construct the kan operon of the kanamycin resistance gene initiated by the lac operator sequence, and replace the wild-type lacI gene and its promoter of Escherichia coli JM109 by homologous recombination. The recombinant plasmid pUC19-tet-PlacIq-lacI, which is a seamless recombination tool gene string composed of a reverse connection of the target gene, uses the tet-PlacIq-lacI tool gene string to replace the natural promoter of the talB gene in Escherichia coli JM109 with the trc promoter ,Proceed as follows:
[0039] 3.1. The plasmid pKD46 was electrotransformed into Escherichia coli JM109, and the strains successfully introduced with the plasmid were obtained by screening with ampicillin plate.
[0040] 3.2. Use the upstream and downstream primers lacI-Plac-kan-F and lacI-kan-R (DNA sequences shown in SEQ ID NO.1 and SEQ ID NO.2 respectively) to obtain Plac-lacO- from plasmid pKD4 by pcr amplification For the kan operon gene, its homologous recombination ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com