Function-specific molecular marker of rice blast resistance gene pi64 and its method and application
A resistance gene and molecular marker technology, applied in the field of agricultural biology, can solve problems such as rice breeding practices that have not yet been applied, and achieve the effects of improving breeding efficiency, wide applicability, and clear breeding selection goals.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Pi64 Amino acid sequence alignment of its alleles and identification of specific amino acid sites
[0047] Through PCR amplification and sequencing methods, from Wool Valley (YMG), Qitou Baigu, 93-11 (Yangdao No. Genomic DNA obtained from Nong 363 and Lijiang Xintuan Heigu (LTH) Pi64 DNA sequence of allele coding region, 3 genes of reference variety Nipponbare obtained from rice database LOC_Os01g57280 , LOC_Os01g57310 and Pish DNA sequence of the coding region. Using the software tool Primer Premier 5 to translate the DNA sequence into an amino acid sequence, and then using the software tool to perform multiple sequence alignment analysis on the obtained amino acid sequence, it was identified that Pi64 The four amino acid sites N698, Y1003, A1031 and V1177 unique to the gene ( figure 1 ).
Embodiment 2
[0048] Example 2: Pi64 Upstream sequence analysis of gene coding region
[0049] Amplified by conventional PCR Pi64 The upstream sequence of the coding region of the gene was sequenced to obtain a 1597bp sequence. Through NCBI BLAST comparison (http: / / blast.ncbi.nlm.nih.gov / Blast.cgi?PROGRAM=blastn&BLAST_ PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome), a 715bp TIR type was found upstream of the coding region transposon, but no homologous sequence of this transposon was found in rice.
Embodiment 3
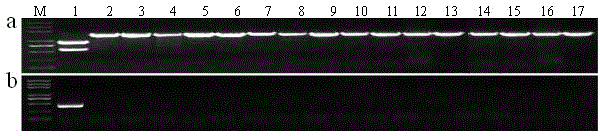
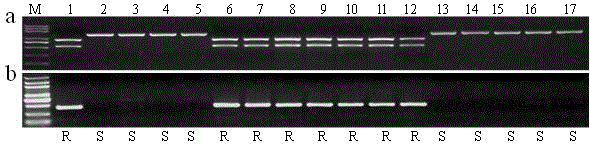
[0050] Example 3: Pi64 Development and detection of gene-specific molecular markers
[0051] use exists in Pi64 The specific amino acid site N698 of the gene ( Pi64 The corresponding codon in the protein is AAT, and the other varieties are GAT), according to the principle of CAPS marker design, using the online software dCAPS Finder 2.0 (http: / / helix.wustl.edu / dcaps / dcaps.html) and Primer-BLAST (http : / / www.ncbi.nlm.nih.gov / tools / primer-blast / index.cgi? LINK_LOC=BlastHome) to design primer YRT3, the nucleotide sequence of the primer pair is shown as SEQ ID NO.1 and SEQ ID NO. 2. The PCR amplification reaction system of YRT3 is: 2 × PCR Buffer for KOD FX 10 μl, dNTPs (2mM each) 4 μl, primer (10pmol, F+R) 1 μl, KOD FX enzyme (1U / μl, TOYOBO) 0.4 μl , DNA template (100ng / μl) 2 μl, add ddH 2 0 to 20 μl. Amplification reaction conditions: 94°C for 2 min; 98°C for 10 s → 59°C for 1.5 min → 68°C for 5 min, 35 cycles; 68°C for 5 min, stored at 10°C. After the PCR reaction, tak...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com