SNP site related to growth trait of paralichthys olivaceus as well as screening method and application of SNP site
A technology of growth traits and screening methods, applied in the field of screening in aquaculture, can solve the problems of long breeding cycle, low efficiency, lack of effective breeding methods for target traits, etc., and achieve the effect of increasing selection intensity and shortening breeding cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
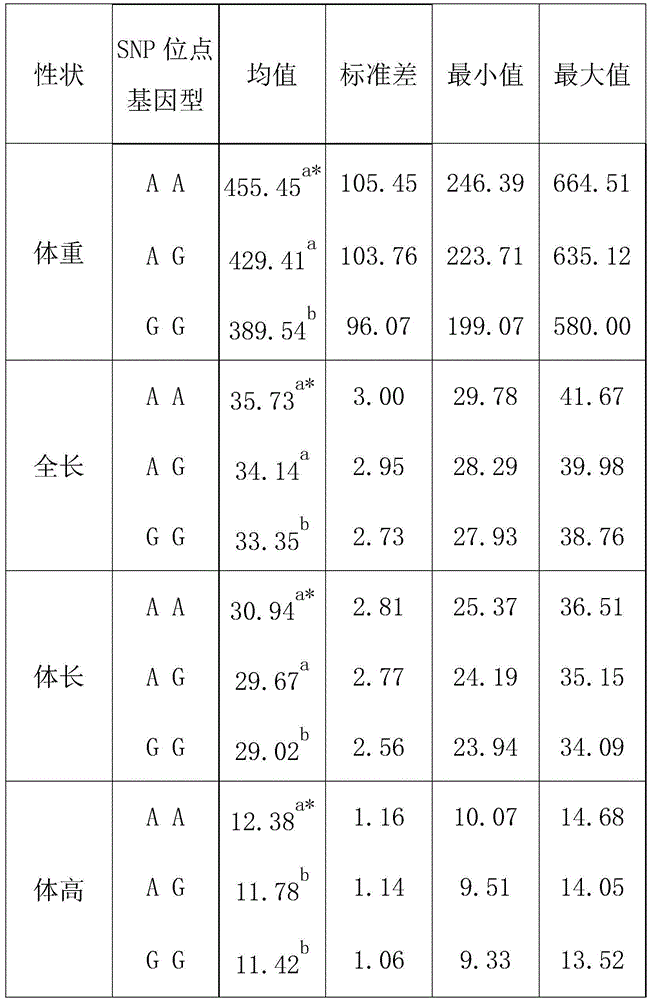
[0030] A SNP site related to the growth traits of flounder, the SNP site is the 33rd bp position of the fourth intron in the GH gene whose sequence is SEQ ID NO: 1, and its base is G or A.
Embodiment 2
[0032] A method for screening SNP sites described in Example 1, the sequence of the upstream primer used is shown in SEQ ID NO: 2; the sequence of the downstream primer is shown in SEQ ID NO: 3.
[0033] Wherein, the sequence of SEQ ID NO: 2 is AGGTTACATCCAAACTGTCAG;
[0034] The sequence of SEQ ID NO: 3 is GTGAGCTGGAGCACCGAACTC.
[0035] In the process of amplifying the GH gene using the upstream and downstream primers, the reaction system used is: total volume 50 μL, DNA 1 μL, 10*buffer 5 μL, Mg 2+5 μL, dNTP 1 μL, primer 1 μL / strip, Taq enzyme 0.6 μL, the balance is water.
[0036] The PCR amplification program used was: pre-denaturation at 94°C for 3 min; then 35 cycles, each cycle including the following steps in turn: denaturation at 94°C for 15 s, annealing at 55°C for 15 s, Extend at 72°C for 30s; after completing 35 cycles, extend at 72°C for 3min.
Embodiment 3
[0038] A method for screening flounder with the SNP site described in Example 1, comprising the following steps:
[0039] A. Screening of flounder: according to the growth traits index in GB / 18654.3-2008 as a standard, different flounders are screened, and the growth traits indexes include but not limited to full length, body length and body height; after the flounder is screened out, the Each flounder was induced to obtain different gynogenetic families and / or heterozygous clonal families, and each gynogenetic family was marked; in this embodiment, 3 different gynogenetic families were selected, and 3 different gynogenetic families were selected. 6 different heterozygous clonal families, fluorescently labeled 6 different families;
[0040] B, polyculture in the same pond: the flounder after marking is polycultured in the same pond for 15 months;
[0041] C. Genotyping: After 15 months of breeding, each family randomly selected 20 tails, a total of 120 tails, and identified t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com