Probe, gene chip and method for detecting expression abundance of circular RNA
A technology of expression abundance and gene chip, applied in the field of detection of circular RNA expression abundance, can solve the problem of low research throughput and achieve the effect of high-throughput detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
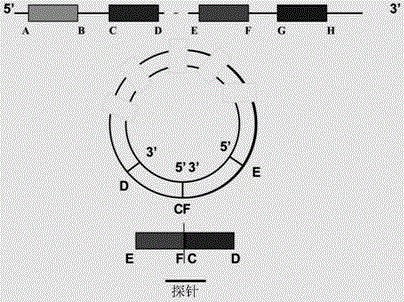
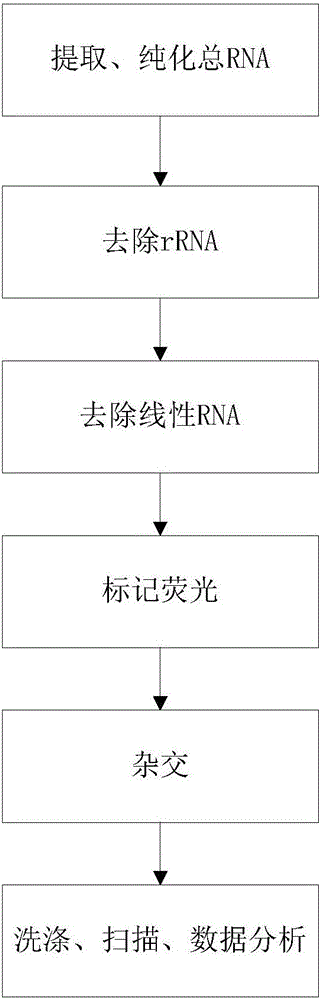
[0028] In order to have a more specific understanding of the technical content, characteristics and efficacy of the present invention, the specific method for detecting the expression abundance of circular RNA of the present invention is described in detail as follows in conjunction with the accompanying drawings:
[0029] 1. Design chip probe sequence
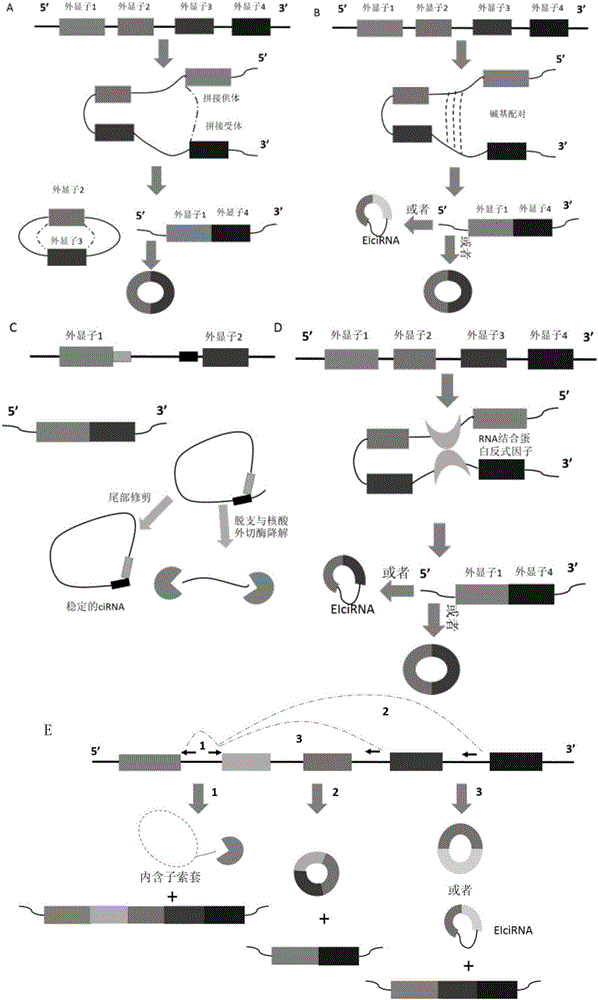
[0030] According to the special splicing pattern of circular RNA-there is a specific reverse splicing site where the 5'end of the donor exon and the 3'end of the acceptor exon are connected end to end (backsplicing, linear RNA does not have this site) , The circular RNA probe is designed on the 60nt chip probe sequence located at the reverse splicing site, which can specifically detect circular RNA in the sample.
[0031] The circular RNA detected in this example is hsa_circ_0034688, which is formed by reverse splicing from the fourth exon to the fifth exon of a gene named OIP5. According to the 5'end sequence of the circular RNA:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com