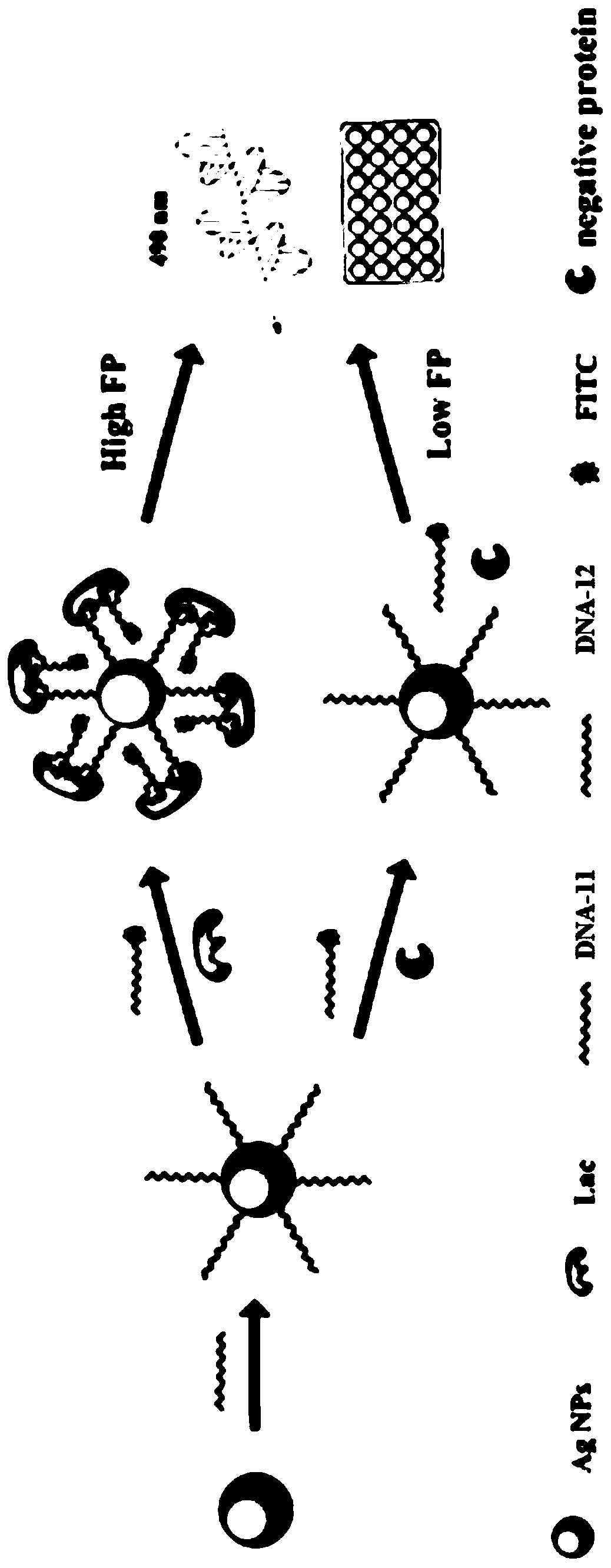
Fluorescent polarization aptasensor based on dual signal amplification and its application
An aptamer sensor, dual-signal amplification technology, applied in fluorescence/phosphorescence, instruments, scientific instruments, etc., can solve problems such as background signal generation and interference detection, and achieve strong anti-interference, high sensitivity, fast and high throughput The effect of detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Analysis of the best aptamer truncation sites
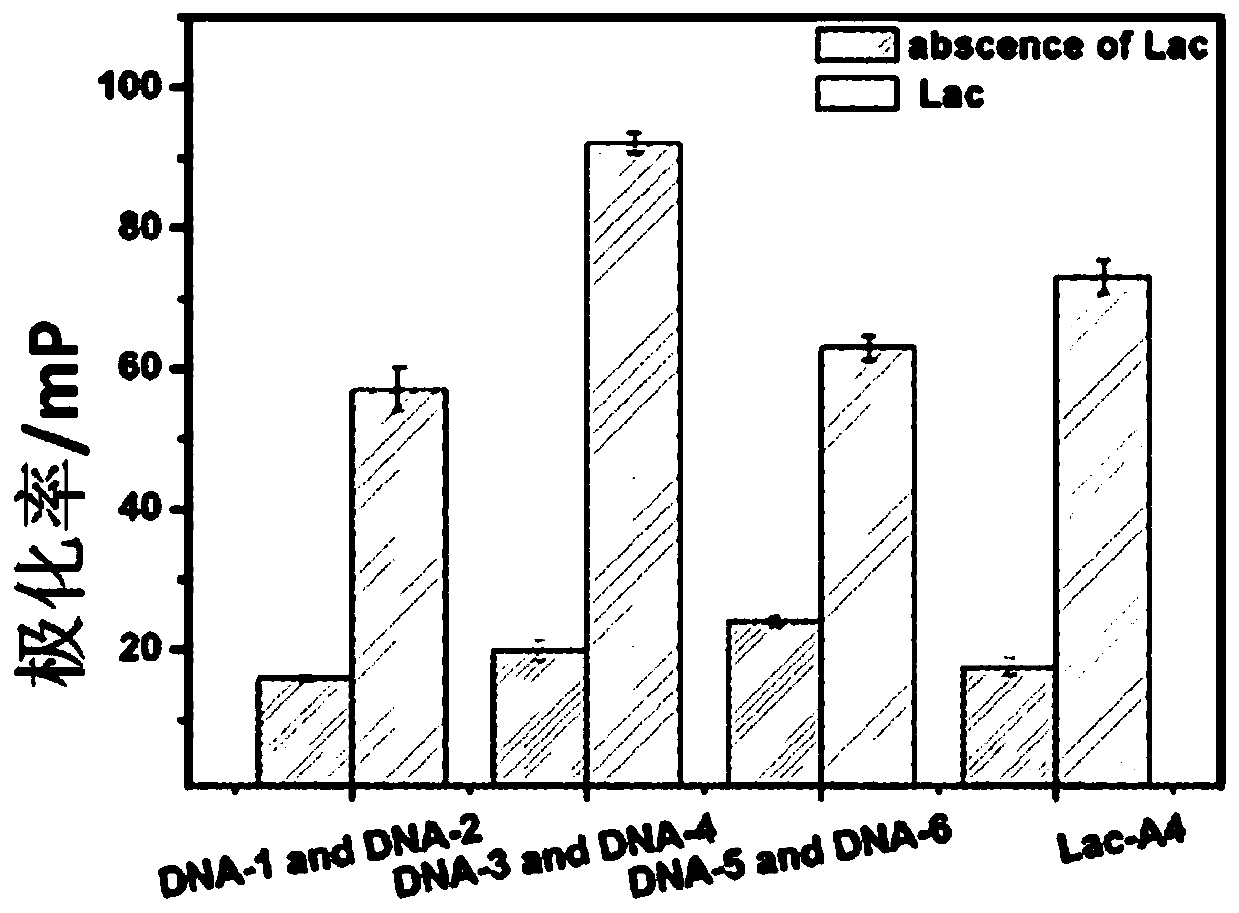
[0044] The previously screened aptamer A4SEQ ID NO.16: AGGCAGGACACCGTAACCGGTGCATCTATGGCTACTAGCTCTTCCTGCCT was truncated at 15, 25, and 35 bases respectively to obtain three groups of truncated aptamers: DNA-1 (15 bases) and DNA-2 (35 bases ), DNA-3 (25 bases) and DNA-4 (25 bases), DNA-5 (35 bases) and DNA-6 (15 bases). DNA-1, DNA-3 and DNA-5 all modify FITC. Mix 10 μL and 250 μM three groups of cut-off aptamers with 1 μg / mL lactoferrin standard sample respectively, react at 37°C for 30 min, detect with a microplate reader, the excitation wavelength is 480 nm, the emission wavelength is 528 nm, and finally the best cut-off aptamer is obtained. Ligand;
[0045] figure 2 Plot of aptamer fluorescence polarization signal truncated for different base numbers. It can be seen from the figure that the signals of DNA-3 and DNA-4 are the highest, and are significantly higher than that of the complete aptamer Lac-A4, a...
Embodiment 2
[0046] Example 2: Optimization of truncated aptamers
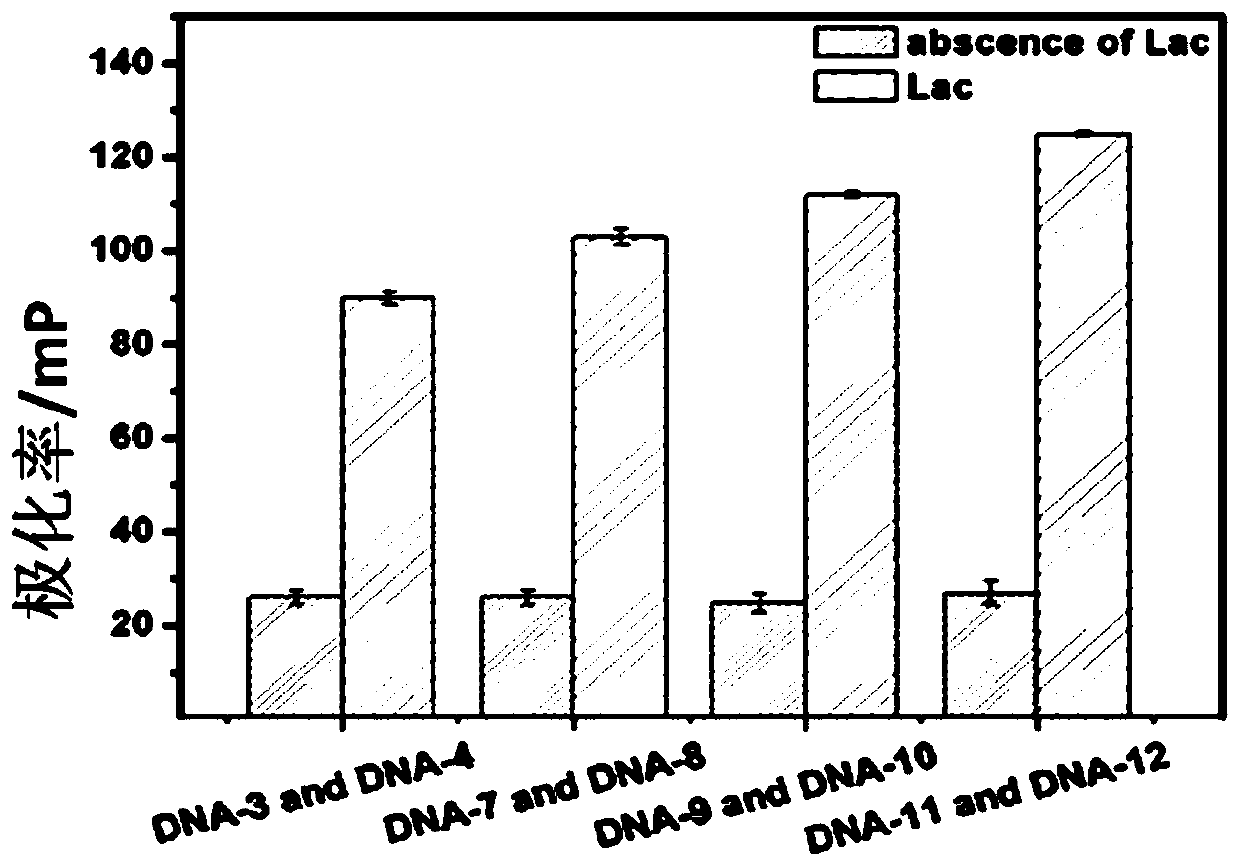
[0047]The obtained truncated aptamer structure was continuously optimized. Three pairs of truncated aptamers: DNA-7 and DNA-8, DNA-9 and DNA-10, DNA-11 and DNA-12 were obtained by removing 3, 5 and 7 hybrid base pairs of the truncated aptamers, respectively. Four groups of 10 μL and 250 μM truncated aptamers were mixed with 90 μL and 1 μg / mL lactoferrin standard samples respectively, reacted at 37°C for 30 min, and detected with a microplate reader.
[0048] image 3 Fluorescence polarization signal plot optimized for truncated aptamer structures. It can be seen from the figure that as the distance between the complementary base pairs of the two truncated aptamers decreases, the fluorescence polarization signal increases gradually, and the signal is the highest when the complementary base pairs are completely removed, and the background also increases with the complementary base The base pair decreased, indicating that ...
Embodiment 3
[0049] Example 3: Dual Binding Site Validation
[0050] Destroy the arm loop structure of the best truncated aptamers DNA-11 and DNA-12 respectively to obtain DNA-13 and DNA-14. Mix 10 μL, 250 μM DNA-13, DNA-14 with 90 μL, 1 μg / mL lactoferrin standard sample , react at 37°C for 30 min, and detect with a microplate reader. Compare the changes in the signal before and after. Afterwards, the two truncated aptamers were labeled with FITC, namely DNA-11 and DNA-15, and the signals generated by them and the signals generated after mixing were compared, and a conclusion was drawn.
[0051] Figure 4 Dual binding site validation fluorescence polarization signal plot. Firstly, when the arm loop structures of DNA-11 and DNA-12 were removed, the signals were significantly reduced, indicating that the arm loop structures of DNA-11 and DNA-12 were very important in binding lactoferrin, and their arm loop structures were The loop structure functions to specifically recognize the protein...
PUM
| Property | Measurement | Unit |
|---|---|---|
| linear range | aaaaa | aaaaa |
| linear range | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com