A method for constructing signal transduction network based on multivariate Granger test
A technology of signal transduction and network construction, applied in informatics, genomics, bioinformatics, etc., can solve problems such as inapplicability, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
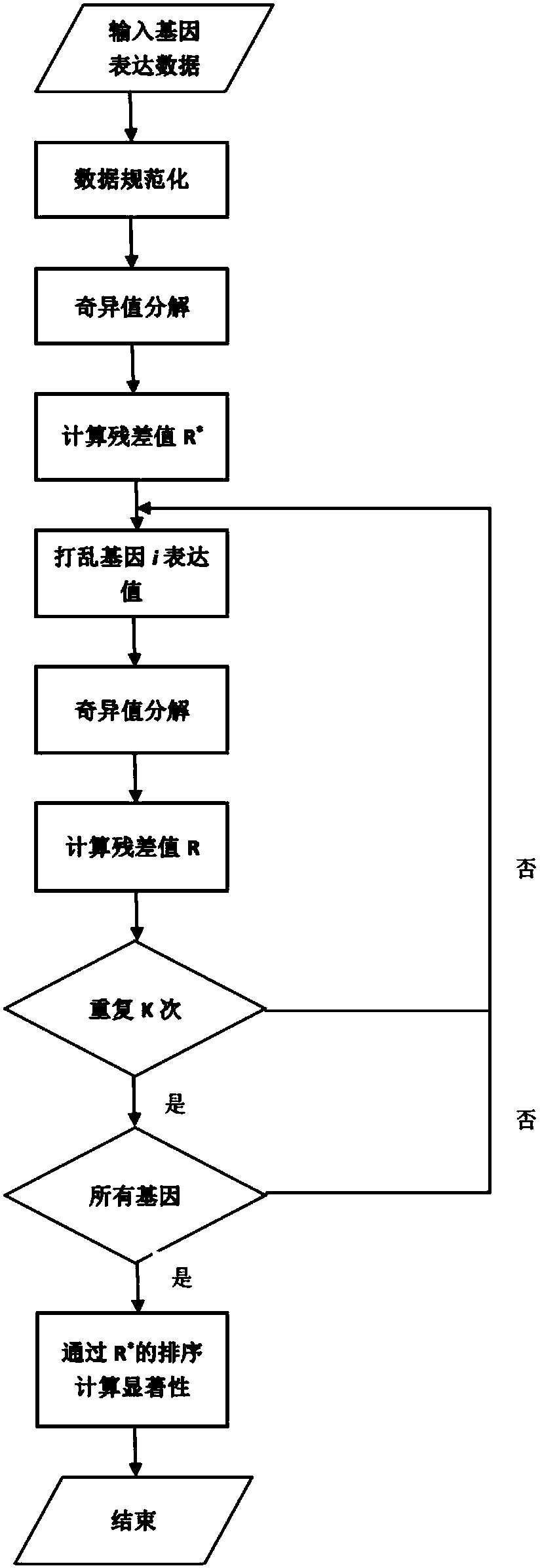
[0028] 1. Preprocessing of gene expression data
[0029] Read in the time series gene table data file with g i, t Represents the expression value of gene i at time t, and performs two-norm normalization under the time series expression group of this gene:
[0030]
[0031] M represents the normalized number of time slices of gene expression data x i, t It is used as the gene expression value in subsequent steps.
[0032] Second, construct the coefficient matrix
[0033] Based on the assumptions of time stationarity and the first-order Markov model, a linear regression equation is constructed:
[0034] x i, t =∑ 0 r j, t-1 x j, t-1 (2)
[0035] Where x j, t-1 Indicates the expression value of gene j at time t-1, r j, t-1 Represents the influence coefficient of the expression value on gene i at time t-1, and N represents the total number of genes. Obtain the matrix equation according to the linear equation:
[0036]
[0037] Where r i, j Indicates the influence coefficient of gene i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com