Thalassemia gene detection method based on high-throughput sequencing technology
A technology for thalassemia and gene detection, applied in the biological field, can solve the problems of inability to detect α&β-thalassaemia at the same time, high price, etc., and achieve the effect of small amount of sample DNA and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1: Establishment of α&β-thalassemia detection method
[0069] (1) Primer design
[0070] Through the Primer Premier 5.0 software, for 6 types of α-thalassemia and 26 mutant types of β-thalassemia, amplified primer pairs were designed. The primer sequences and pairings are as follows:
[0071] (1) Six types of amplification primers for α-thalassaemia:
[0072] Sea-F: 5'-GAGGGAAGGAGGGGAGAAGCTGAGT-3'
[0073] Sea-R: 5'-AGCCCACGTTGTGTTCATGGC-3'
[0074] 3.7-F: 5'-CCCCACATCCCCTCACCTACATTC-3'
[0075] 3.7-R: 5'-GCATCCTCAAAGCACTCTAGGGTCC-3'
[0076] 4.2-F: 5'-TGCTTTTGTGAGTGCTGTGTTGACC-3'
[0077] 4.2-R: 5'-GAAGTAGCTCCGACCAGCTTAGCAAT-3'
[0078] α2R: 5'-CGGGCAGGAGGAACGGCTAC-3'
[0079] Among the above primers, Sea-F and Sea-R are amplification-α SEA Forward and reverse primers of the gene fragment; 3.7-F and 3.7-R are amplification-α 3.7 Forward and reverse primers of the gene fragment; 4.2-F and 4.2-R are amplification-α 4.2 Forward and reverse primers of the...
Embodiment 2
[0130] Example 2: Detection of α & β-thalassemia gene mutation types in clinical samples
Embodiment 3
[0137] Example 3: Detection of clinical samples of α-thalassemia deletion type
[0138] Samples 17, 18, 19, 20, 21, 22, 23 and 24 were detected by the method of Example 1.
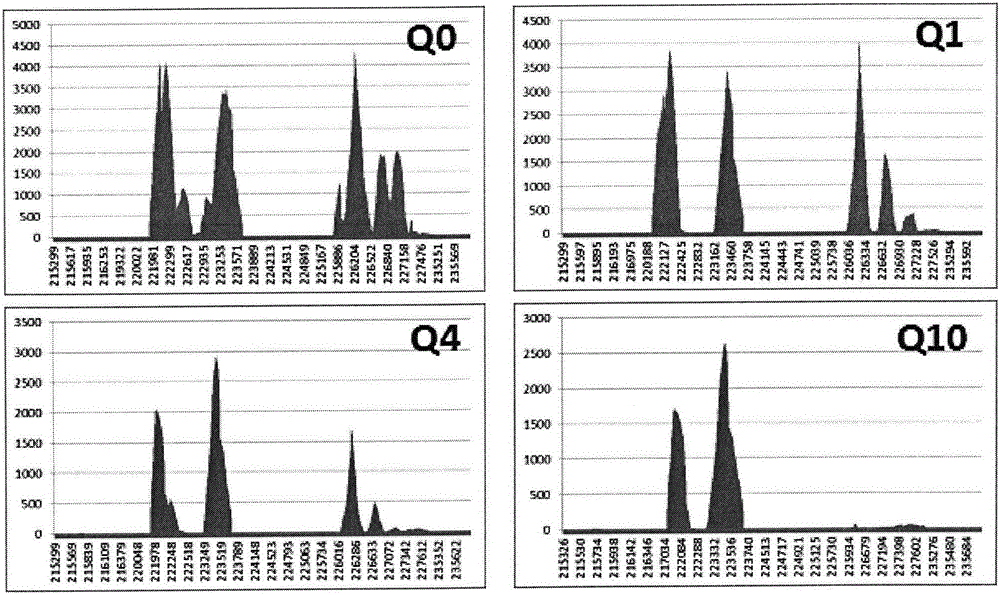
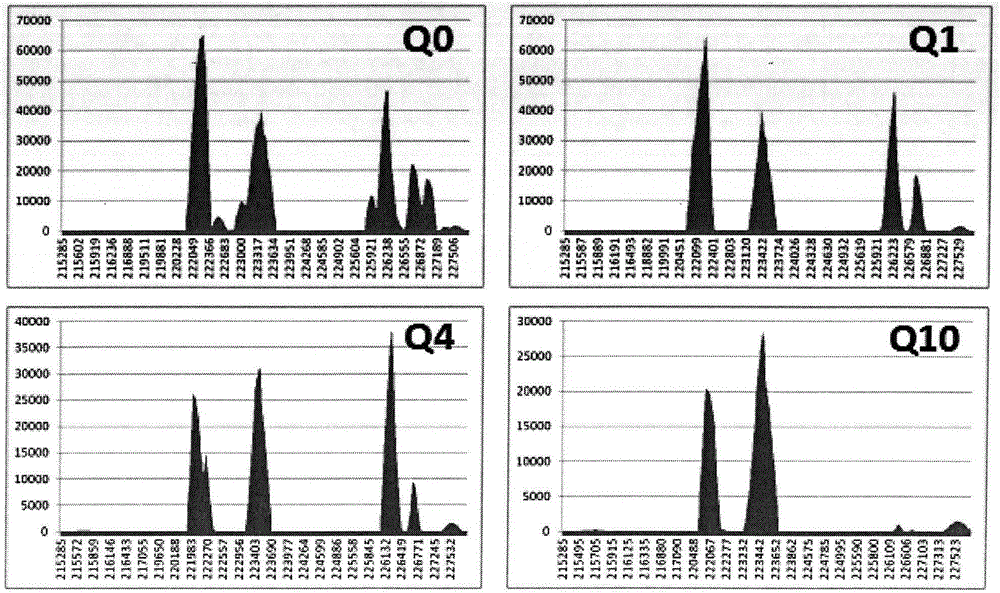
[0139] According to the plug-in established by the method provided by the present invention, the mutation analysis of samples 17, 18, 19, 20, 21, 22, 23 and 24 is carried out, and the mutation frequency is all 0-5%, so samples 17, 18, 19, 20 , 21, 22, 23 and 24 mutation types are all mutation negative, according to the mutation results and Figure 18 , 19 , 20, 21, 22, 23, 24 and 25, the types of samples 17, 18, 19, 20, 21, 22, 23 and 24 are as follows:
[0140]
[0141] The results show that the clinical sample detection of the α-thalassemia deletion type of the present invention is consistent with the original type.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com