A method for detecting alterations in low-abundance molecules
A sample and primer pair technology, applied in the fields of biotechnology and medicine, can solve problems such as the inability to achieve high-sensitivity detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] 1. Primers: The inventor found a pair of primers through a large number of exploratory experiments KRAS Specific primer pair for gene codon 4-14 mutation KRAS -76-F / KRAS -76-R; the sequence is as follows:
[0040] KRAS -76-F: AGGCCTGCTGAAAATGACTG (shown in SEQ ID NO: 1);
[0041] KRAS -76-R: GCAAGAGTGCCTTGACGATA (shown in SEQ ID NO: 2);
[0042] 2. Test method: firstly, the inventors verified the mixture of plasmids with different ratios of mutation ratios to see whether the mutation rate of 0.1% could be detected as stated in the literature. The detection sensitivity of the method according to the invention was determined.
[0043] 2.1. Use plasmid standards (100% mutant plasmid and 100% wild type plasmid) to configure mixed plasmids with different mutation ratios, 100% mutation, 90% mutation, 80% mutation, 70% mutation, 60% mutation, 50% mutation , 40% mutation, 30% mutation, 20% mutation, 10% mutation, 5% mutation, 2.5% mutation, 1% mutation, 0.5% mutation, 0...
Embodiment 2
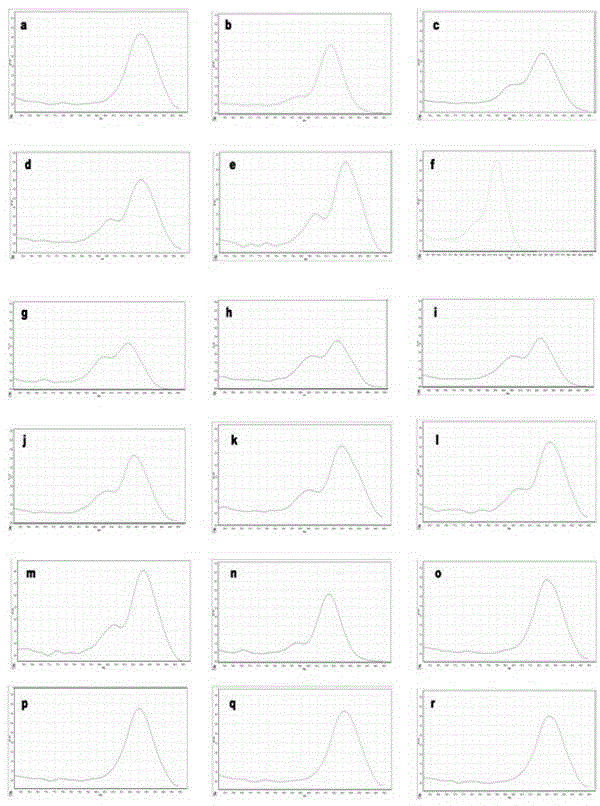
[0053] Utilize the specific primer pair of the present invention KRAS -76-F / KRAS -76-R, verification of digitalHRM method detection results and sequencing method detection results of DNA templates of cancer specimens from different sources (gastric cancer, colon cancer, rectal cancer, esophageal cancer, pelvic cancer, pancreatic cancer, anal canal cancer, breast cancer, lung cancer) consistency rate.
[0054] The specific operation is as follows: select the DNA of paraffin specimens of different diseases as shown in Table 1 below for digitalHRM detection, and use the sequencing method to verify the detection results. The experimental method, experimental apparatus and relevant reaction conditions are as shown in Example 1.
[0055] Table 1
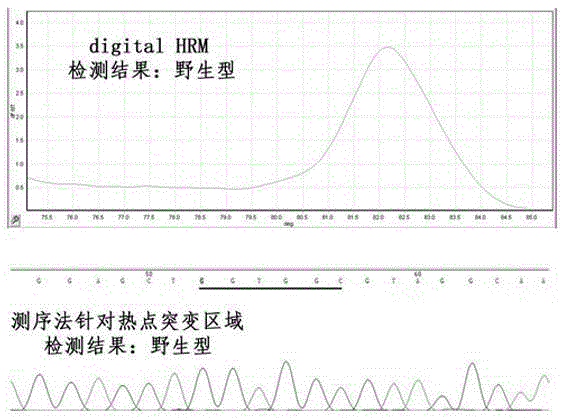
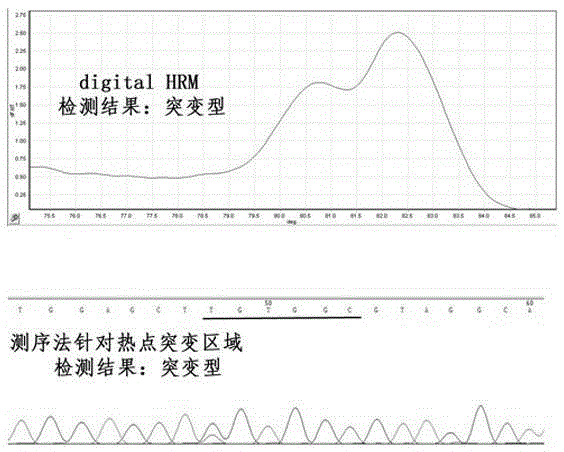
[0056] sample number pathological diagnosis Sequencing results digital HRM results Remark A1 stomach cancer Wild type Wild type See figure 2 shown A2 colon cancer GGT-GAT mutant See image 3 ...
Embodiment 3
[0059]DNA samples with severe fragmentation and poor quality are selected and detected using the method of the present invention. The experimental method, experimental apparatus and relevant reaction conditions are as shown in Example 1. Poor quality DNA samples are defined as: samples whose initial DNA concentration is lower than 50ng / μl, or the ratio of ultraviolet absorption A260 / A280 is lower than 1.6 or greater than 2.0. Select the samples described in Table 2 according to the above requirements.
[0060] Table 2
[0061] Sample serial number Concentration (ng / μl) Purity (A260 / 280) B1 32.6 1.343 B2 33 1.365 B3 23.3 1.245 B4 52.6 1.623 B5 54.9 2.598 B6 203.9 2.118
[0062] While using the method described in Example 1 of the present invention to detect the mutation status of the samples in Table 2, the present invention also uses the common PCR method to detect the samples in Table 2. The common PCR reaction system an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com