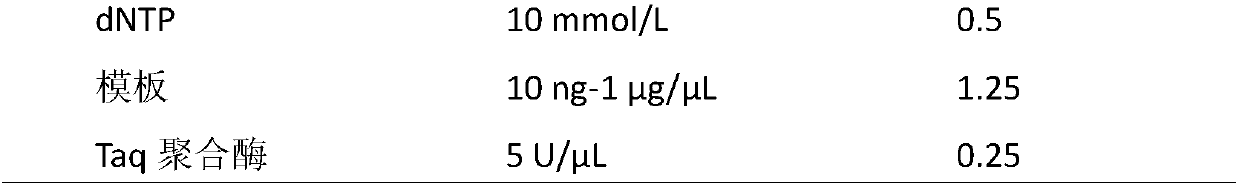A kind of method and special-purpose dna bar code fragment of the identification method of Pleurotus eryngii strain
A barcode, eryngium eryngii technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of unreliable results and time-consuming, improve the identification accuracy and shorten the identification time Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1, the establishment of the acquisition of DNA barcode fragment and method for identification of Pleurotus eryngii strains
[0038] 1. Genomic DNA acquisition of Pleurotus eryngii strains
[0039] Genomic DNA of Pleurotus eryngii strains was extracted.
[0040] 2. PCR amplification
[0041] Using the genomic DNA obtained in 1 above as a template, PCR amplification was performed with primer 728F and primer 1567R.
[0042] 728F: 5'-CATCGAGAAGTTCGAGAAGG-3' (SEQ ID NO: 1)
[0043] 1567R: 5'-ACHGTRCCRATACCACCRATCTT-3' (SEQ ID NO: 2)
[0044] The above-mentioned PCR amplification system is as follows in Table 1:
[0045] Table 1 is the composition of the PCR reaction system (25 μL)
[0046]
[0047] The above PCR amplification program is: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 65°C for 55 s, decreasing 1°C for each cycle, extension at 72°C for 90 s, 10 cycles; denaturation at 94°C for 30 s, annealing at 55°C for 55 s...
Embodiment 2
[0053] Embodiment 2, distinguish Pleurotus eryngii bacterial classification
[0054] 1. Specific detection by the method of the present invention
[0055] The genomic DNAs of known varieties of Pleurotus eryngii strains and Pleurotus abalonus strains were extracted respectively.
[0056] Using the genomic DNA of each variety as a template, the above-mentioned primers 728F and 1567R were used to amplify to obtain PCR products of each variety.
[0057] The PCR product of each kind is sent for sequencing, and the results are as follows:
[0058] The nucleotide sequence of the PCR product of the known varieties of Pleurotus eryngii strains is sequence 3 (DNA barcode fragment for identification of Pleurotus eryngii).
[0059] The nucleotide sequence of the PCR product of known varieties of oyster mushroom strains is sequence 4 (the 1st-20th position of the sequence 4 is the upstream primer 728F, and the 385th-407th position of the sequence 4 is the reverse complementary sequence ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com