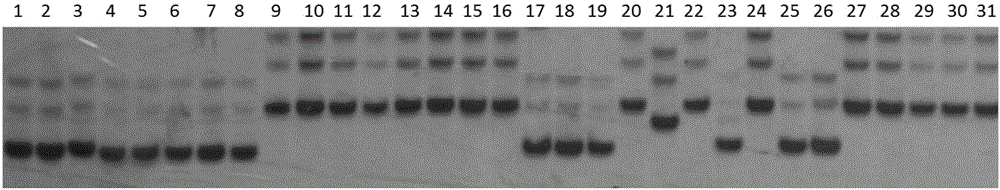
SSR (simple sequence repeat) primer pair linked to fruiting rate on rice chromosome 2 and application of SSR primer pair
A primer pair and seed setting rate technology, applied in the field of plant molecular genetics, achieves the effects of low cost, clear directionality, and simple identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 : Primer Screening
[0023] SSR primers were downloaded from www.genomene.org; the sequences of RM341SSR primers are shown in Table 1. The sequences of the SSRs in Table 6 themselves were obtained from publicly available web resources.
[0024] Table 1 RM341 sequence
[0025]
Embodiment 2
[0026] Example 2 : The CTAB method extracts the total DNA of rice, the steps are as follows:
[0027] (1) Weigh 0.4-0.5g of fresh leaves for each material, add liquid nitrogen to the mortar, grind and pulverize quickly, and then transfer to a 2.0mL sterile centrifuge tube; before this, dry materials are pulverized with a pulverizer , and take the powder;
[0028] (2) Add 700 μL of preheated 65°C CTAB extraction buffer (pH 8.0) to each tube, bathe in 65°C water for 45 minutes, take out and shake well once every 5 minutes;
[0029] (3) Add 700 μL of chloroform / isoamyl alcohol (24:1) and shake gently for 8 minutes, then centrifuge at 12000 r / min for 10 minutes;
[0030] (4) Take the supernatant and transfer it to another 1.5 mL sterile centrifuge tube, add 600 μL of pre-cooled isopropanol, mix it upside down, and freeze it in a -20°C refrigerator for more than 30 minutes;
[0031] (5) centrifuge the centrifuge tube at 12000r / min for 10min, and take the precipitate;
[0032] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com