A method of performing targeted editing on a mitochondrial genome by utilizing CRISPR/Cas9
A mitochondrial genome and mitochondrial gene technology, applied in the direction of viruses/bacteriophages, biochemical equipment and methods, and the use of vectors to introduce foreign genetic materials, can solve problems such as cell growth stagnation, energy production hindrance, and protein translation inhibition.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
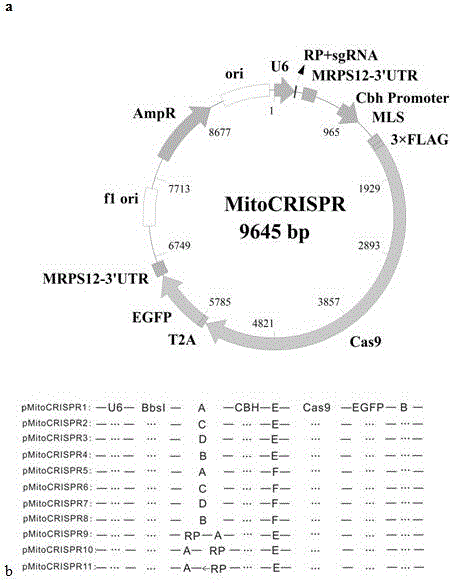
[0050] Example 1 Construction of MitoCRISPR plasmid vector
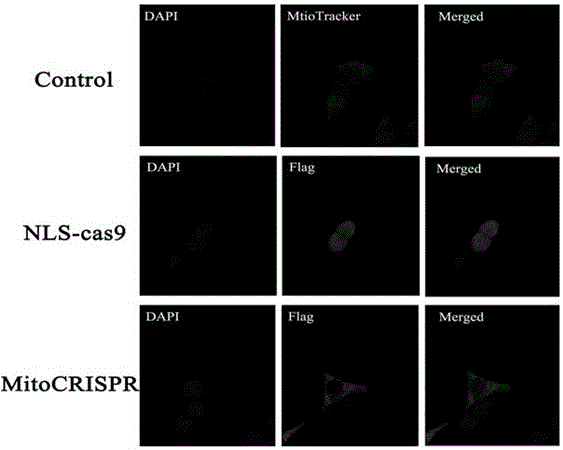
[0051] First, we added the 3'UTR signal of the mitochondrial localization signal MRPS12 gene after the U6 promoter. The plasmid of Mito-U6-RP-BbsI-3'UTR was constructed. Then remove the nuclear localization signal before the Cas9 gene, add the mitochondrial localization signal of Cox8A, that is, the MLS sequence, and construct the plasmid of Mito-U6-RP-BbsI-3'UTR-CBh-MLS-Cas9. Then add the EGFP gene after Cas9 to observe the transfection efficiency. At the same time, the 3'UTR signal of MRPS12 was added after the Cas9 expression frame to stabilize the entire structure of the expressed Cas9 mRNA. Finally, the Mito-U6-RP-BbsI-3'UTR-CBh-MLS-Cas9-2A-GFP-3'UTR (MitoCRISPR) plasmid vector, namely pMitoCRISPR1 ( figure 1 a).
[0052] Specifically:
[0053] (1) The introduction of elements used to control the expression of sgRNA: After the U6 promoter of the PX459 vector, the mitochondrial localization signal RP sequenc...
Embodiment 2
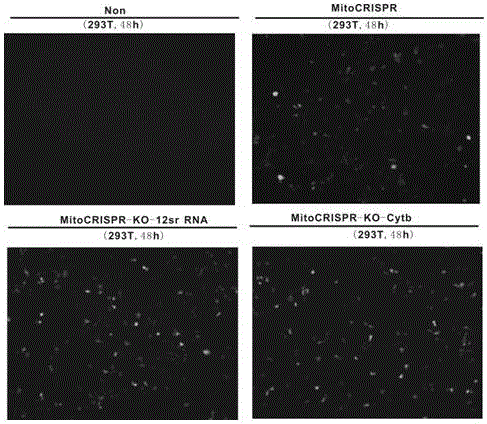
[0063] Example 2 Gene Knockout for the Mitochondrial 12sr RNA Gene Locus
[0064] The specific method for constructing a mitochondrial gene knockout vector targeting the mitochondrial 12srRNA gene locus is as follows:
[0065] Select the pMitoCRISPR1 plasmid as the backbone to construct the mitochondrial gene knockout recombinant plasmid vector. The pMitoCRISPR1 plasmid contains the basic elements of the CRISPR system, namely the Cas9 protein gene and the elements that control the expression of sgRNA in mitochondria, including the U6 promoter, RP sequence and 3'UTR sequence . The U6 promoter is used to promote the expression of sgRNA. The 3'UTR sequence helps stabilize the expressed sgRNA so that it can come out of the nucleus and locate on the mitochondrial outer membrane; the input sequence at the 5' end of the RNA-RP sequence helps the RNA enter the mitochondria. Once the 3'UTR sequence helps RNAs appear on the mitochondrial surface, a second trafficking sequence (RP seque...
Embodiment 3
[0080] Example 3 Mitochondrial gene knockout targeting the mitochondrial cytb gene locus
[0081] In this example, the cytb gene on the mitochondrial genome is selected and knocked out, and the specific steps are as follows: the annealing reaction is performed using the method described in Example 2.
[0082] The sgRNA target sequence of Cytb is: ATCCCGTTTCGTGCAAGAAT; the RP sequence is: TCTCCCTGAGCTTCAGGGAG; the Cytb gene sgRNA primer sequence for insertion into the Bbs I restriction site of pMitoCRISPR1 plasmid is as follows: Mito-KO-H-Cytb-F: CACCGTCTCCCCTGAGCTTCAGGGAGATCCCGTTTCGTGCAAGAAT;
[0083] Mito-KO-H-Cytb-R:AAACATTCTTGCACGAAACGGGATCTCCCTGAAGCTCAGGGAGAC;
[0084] The mitochondrial Cytb gene knockout vector pMitoCRISPR1-KO-Cytb was constructed using the method described in Example 2, and verified by sequencing. Next, the plasmid vector was transfected into 293T cells by the method described in Example 2, and after 3 days, the efficiency of the plasmid vector for mito...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com