Small molecule biochemical profiling of individual subjects for disease diagnosis and health assessment
A technology of small molecules and subjects, applied in disease diagnosis, biomaterial analysis, analytical materials, etc., can solve problems that are not widely used in diagnosing individual patient diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
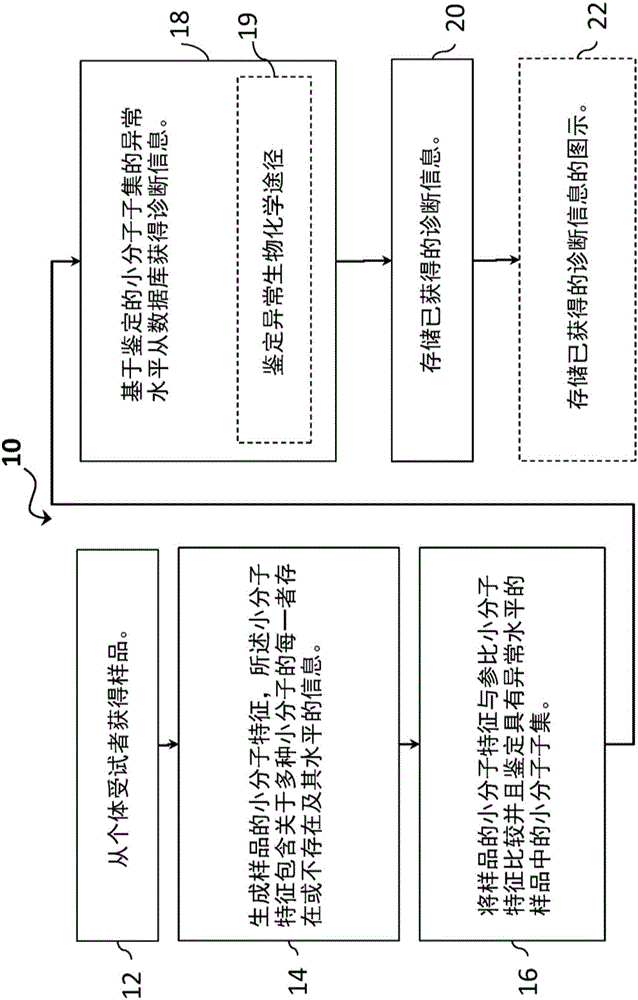
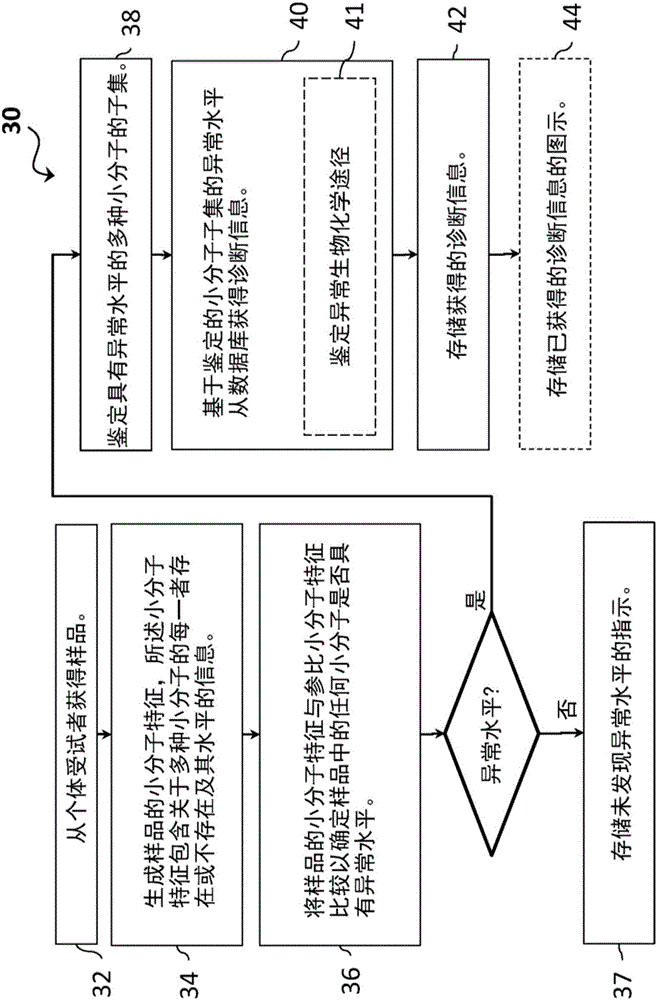
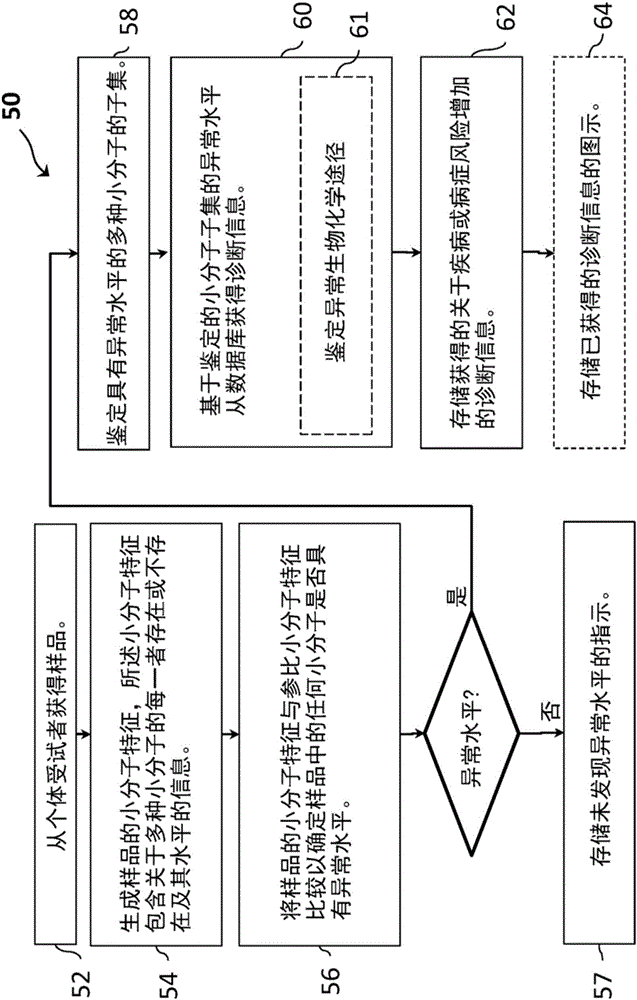
[0076] In one embodiment, there is provided a method of diagnosing or aiding in the diagnosis of a disease or condition in a symptomatic subject by investigating a biological sample from said subject by statistical analysis for endogenous small molecules, The presence, absence, or abnormal levels of dietary small molecules, microbial small molecules, and xenobiotic small molecules to identify outliers for the small molecules that suggest that small molecules are abnormal in the sample, and list abnormal small molecules.
[0077] In another embodiment, abnormal small molecules are visualized. The visualization can display indicators for each of the pathways, including indicators of rare or missing values for the pathways, showing each pathway (eg, super pathway or sub pathway) as normal / common or abnormal. Any number of pathways can be listed in the display, and any number of graphical representations can be used to present the results. In one embodiment, the graphical illu...
Embodiment 1
[0413] Example 1: Evaluation of diseased individuals in a pilot study.
[0414] To evaluate the ability to diagnose or aid in the diagnosis of disease, the method was performed on a cohort of 100 symptomatic individuals, 56 of whom had a known diagnosis (as a "positive control" for the method, N=56) and of whom 44 had an as yet unascertained diagnosis Results ("test" subjects, N=44). Plasma samples were obtained from each individual and small molecules were extracted from aliquots (typically 50-100uL) of the plasma samples. An aliquot of each sample was surveyed across a biochemical library of more than 7000 biochemicals to detect the biochemicals present in the sample. All samples were blinded for diagnosis at the time of analysis. A total of 923 biochemicals were detected, 523 named and 400 unknown. Named biochemicals represent molecules for which reliable chemical standards are available, and represent those for which reliable chemical standards have been analyzed on an ...
Embodiment 2
[0421] Example 2: Clinical Evaluation of Affected Individuals in an Expanded Study
[0422] In another embodiment, the methods described herein are used to evaluate plasma samples from 200 individuals. The 200 samples consisted of the 100 sample cohort described in Example 1 plus 100 samples from a new cohort of symptomatic individuals. All 200 samples included in this study had been analyzed using one or more targeted analyte combinations in quantitative assays performed in clinical diagnostic laboratories. The most common directional tests completed were combination assays of amino acids or acylcarnitines (108 and 26 patients, respectively). Of the 200 subjects in the pooled cohort, 132 subjects were positively diagnosed with an inborn error of metabolism (IEM) by the treating physician; for 68 subjects, a clinical diagnosis could not be determined.
[0423] Using the OrbiElite mass spectrometer in an LC-MS analytical method, each sample is surveyed across the complete lib...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com