Kit for detecting nasopharyngeal cancer susceptibility and SNP marker of kit
A technology of susceptibility and markers, applied in the field of genetic testing, can solve the problems of lack of system integration, improper sampling location, limited detection range, etc., and achieve the effect of good technical reproducibility, high practicability, and high cost performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Used to detect SNP sites associated with susceptibility to nasopharyngeal carcinoma
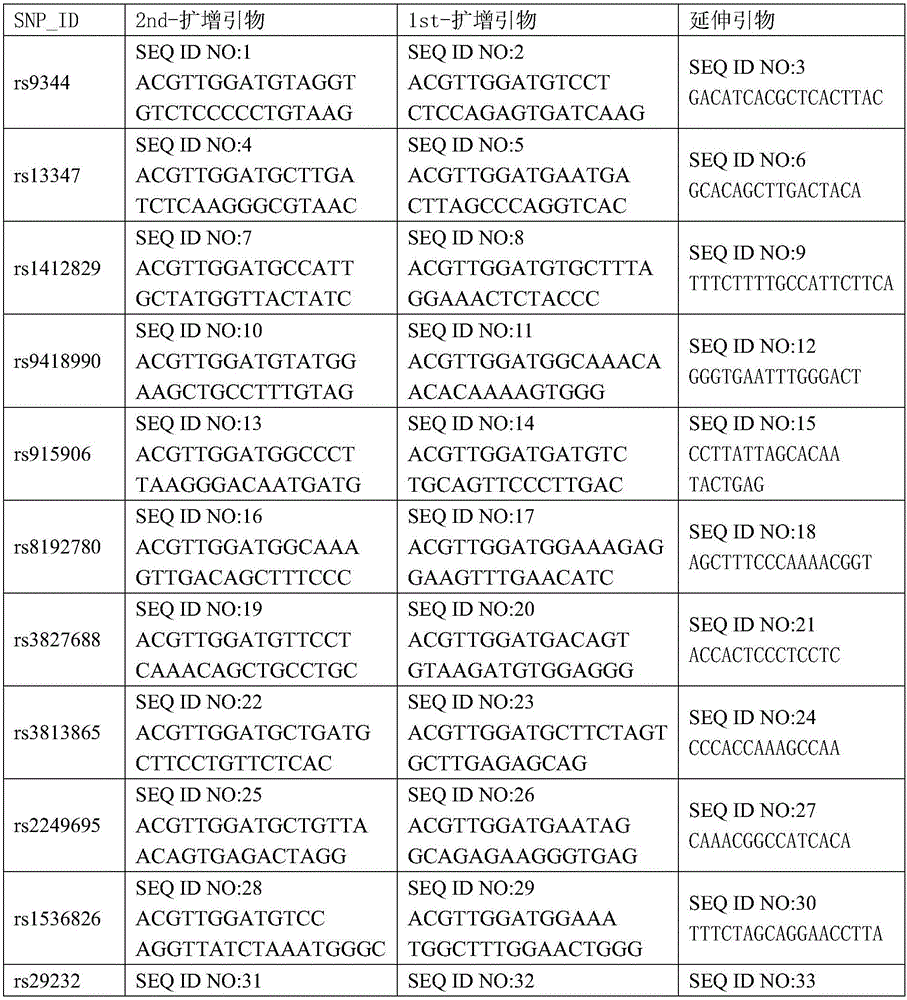
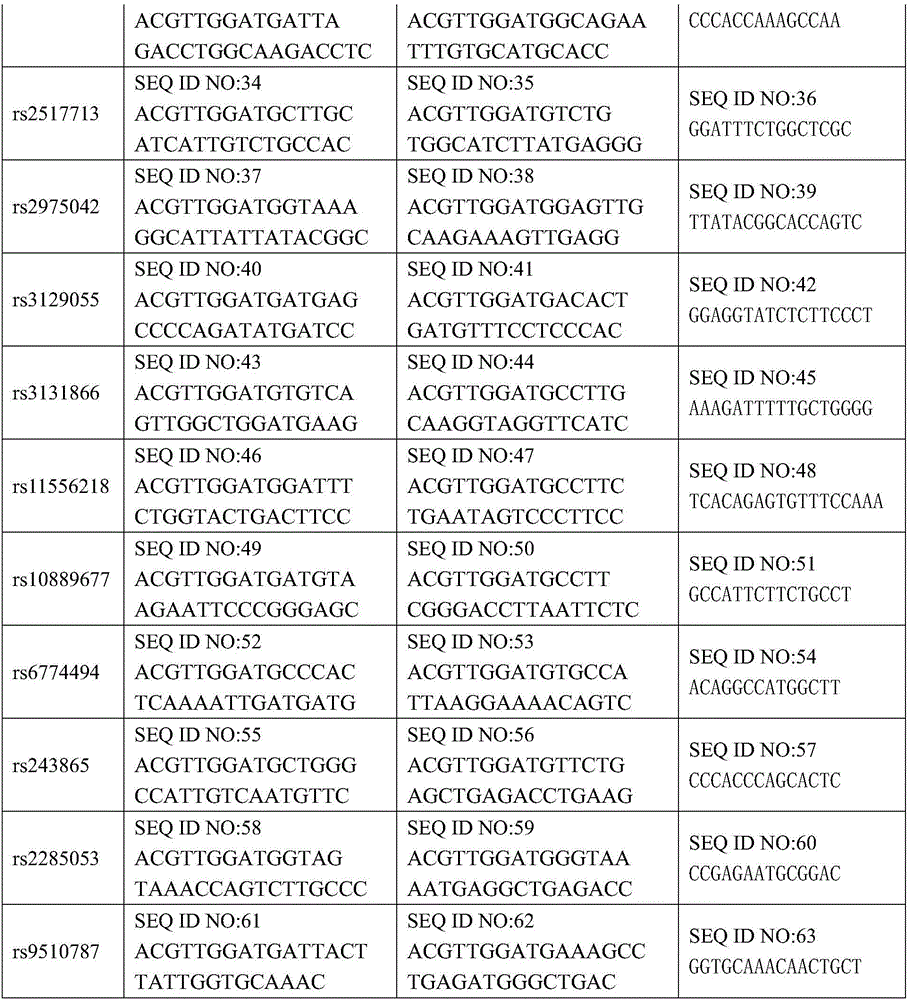
[0039] 其中rs9344位于基因CCND1区域;rs13347位于基因CD44区域;rs1412829位于基因CDKN2A区域;rs9418990、rs915906、rs8192780、rs3827688、rs3813865、rs2249695和rs1536826位于基因CYP2E1区域;rs29232位于基因GABBR1区域;rs2517713和rs2975042位于基因HLA- A region; rs3129055 and rs3131866 are located in the gene HLA-F region; rs11556218 is located in the gene IL16 region; rs10889677 is located in the gene IL23R region; rs6774494 is located in the gene MDS1-EVI1 region; rs243865 and rs2285053 are located in the gene MMP2 region; rs9510787 is located in the gene TNFRSF19 region.
Embodiment 2
[0040] Example 2 Kit for detecting susceptibility to nasopharyngeal carcinoma
[0041] 1. Preparation of the kit:
[0042] 1. Design and synthesize PCR amplification primers and single-base extension primers for the 21 SNP sites. The PCR amplification primers and single base extension primers of the SNP sites to be tested are shown in Table 1
[0043] Table 1 PCR amplification primers and single-base extension primers for SNP sites to be tested
[0044]
[0045]
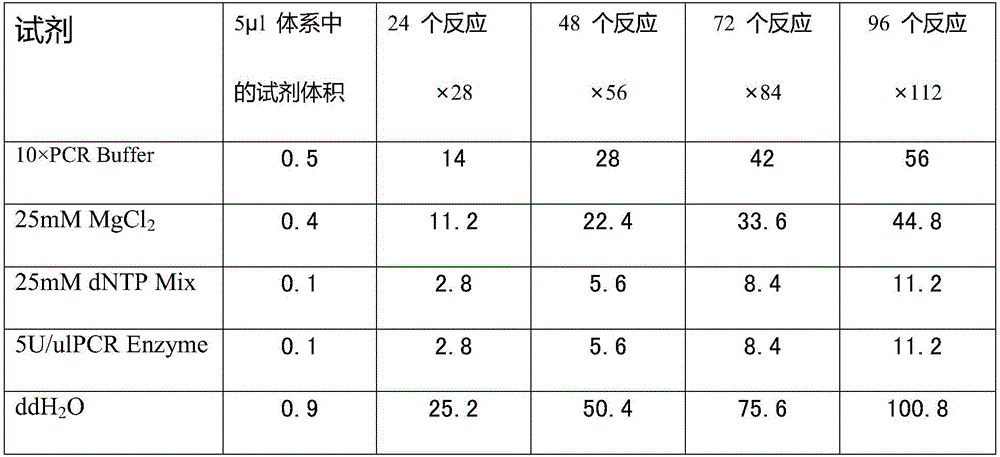
[0046] 2. The kit also includes Taq enzyme, dNTP mixture, MgCl 2 Solution, PCR reaction buffer, enzyme digestion reaction buffer, deionized water.
[0047] 2. The detection method of the kit:
[0048] 1. Extraction of DNA
[0049] 1.1 DNA extraction from oral swab
[0050] 1) Put the oral swab in a 2ml centrifuge tube and add 400μl PBS.
[0051] 2) Add 20 μl QIAGEN Protease and 400 μl Buffer AL. Immediately vortex for 15s to mix. To ensure efficient lysis, sample and Buffer AL must be mixed immediately...
Embodiment 3
[0097] Example 3 The literature cited by 21 SNP sites closely related to nasopharyngeal carcinoma, see Table 5
[0098] Table 5 Literature cited by 21 SNP sites closely related to nasopharyngeal carcinoma
[0099]
[0100]
[0101]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com