DNA sequence k-mer frequency statistical method based on MapReduce
A DNA sequence and statistical method technology, applied in the field of bioinformatics, can solve problems such as the large proportion of I/O overhead in the total processing time, excessive computer performance requirements, and unsatisfactory processing efficiency, etc., to reduce I/O overhead and network transmission overhead, shorten processing time, and reduce the effect of calculation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] The technical solutions in the embodiments of the present invention will be described clearly and in detail below with reference to the drawings in the embodiments of the present invention. The described embodiments are only some of the embodiments of the invention.
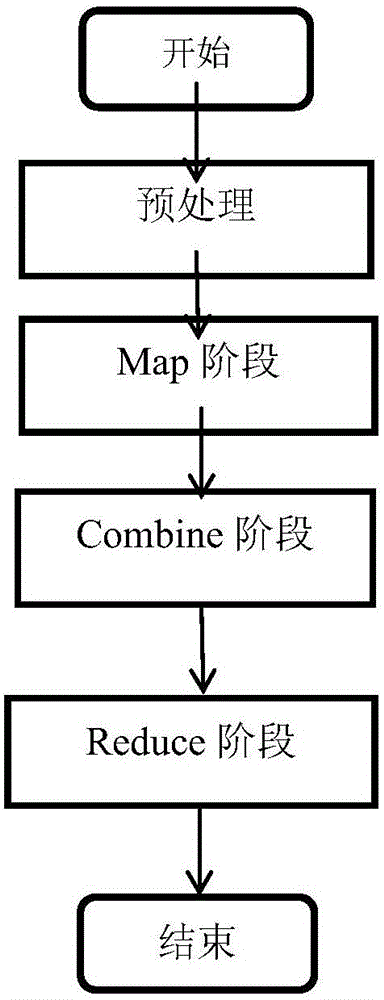
[0042] Such as figure 1 Shown is the main flow diagram of the method of the present invention, and its steps mainly include:
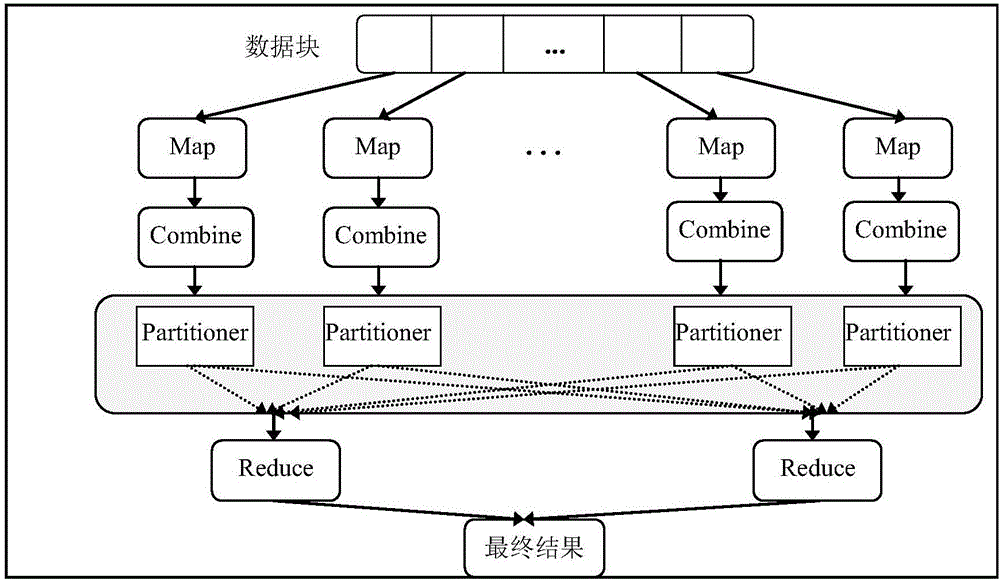
[0043] Step 1: Preprocessing stage. Receive the DNA sequence file that needs to be processed and the variation range parameter of k in k-mer input by the user, and the initial value is set to k 1 , the final value is set to k 2 , with k 1 ≤k≤k 2 . First, the cluster environment running the MapReduce parallel computing model automatically cuts the input DNA sequence files into data blocks of a certain size, and distributes them equally to each node. Then, each node processes the sequence files assigned to the node to remove error sequences and non-DNA coding sequences. The s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com