A method for improving dna cloning and assembly efficiency by using short-chain primer suppression
A short-chain, purpose-built technology that can be used in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc., and can solve problems such as assembly efficiency that cannot be ignored
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0125] Example 1. A method for improving DNA cloning and assembly efficiency by using short-chain primer suppression
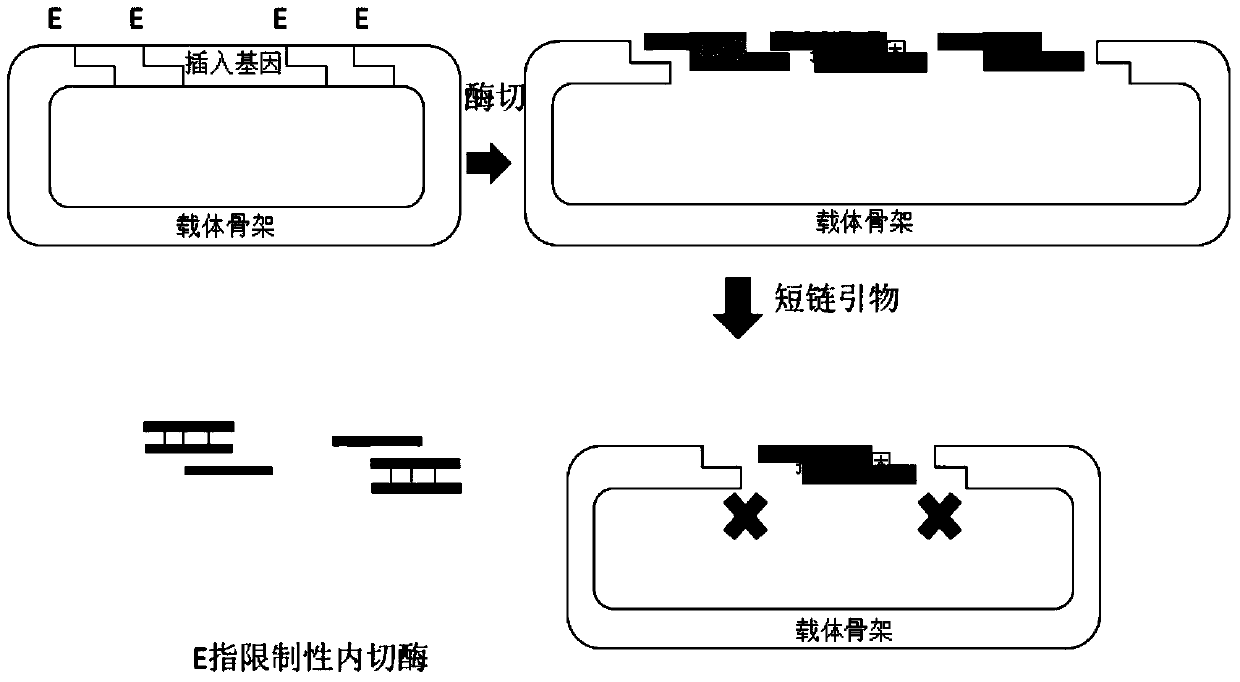
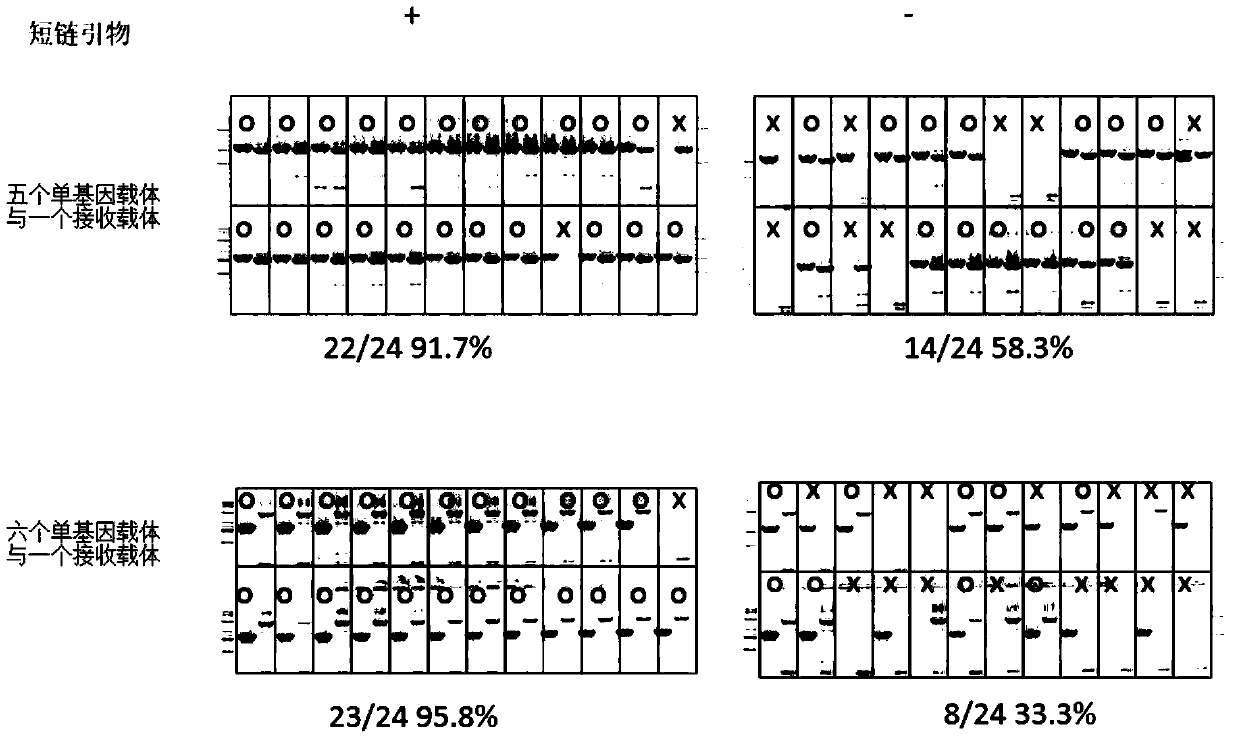
[0126] The specific test process in this embodiment is as attached figure 2 shown.
[0127] 1. Preparation of 7 circular recombination vectors and receiving vector x
[0128] Circular recombination vector 1 includes DNA fragment A 1, DNA fragment 1 to be spliced, DNA fragment B 1 and backbone carrier from upstream (the nucleotide sequence of the backbone carrier is after removing nucleotides 4799 to 5663 in sequence 1 The obtained sequence); the DNA fragment A1 in the circular recombination vector 1 is the 23rd-55th position in the sequence 11, and the DNA fragment B1 is the 395th-426th position in the sequence 11;; the circular recombination vector 1 The nucleotide sequence of the DNA fragment A 1 that is identical or complementary to the short-chain primer A is the 30th-51st position in the sequence 11, the nucleotide sequence of the DNA fragment 1 to be ...
Embodiment 2
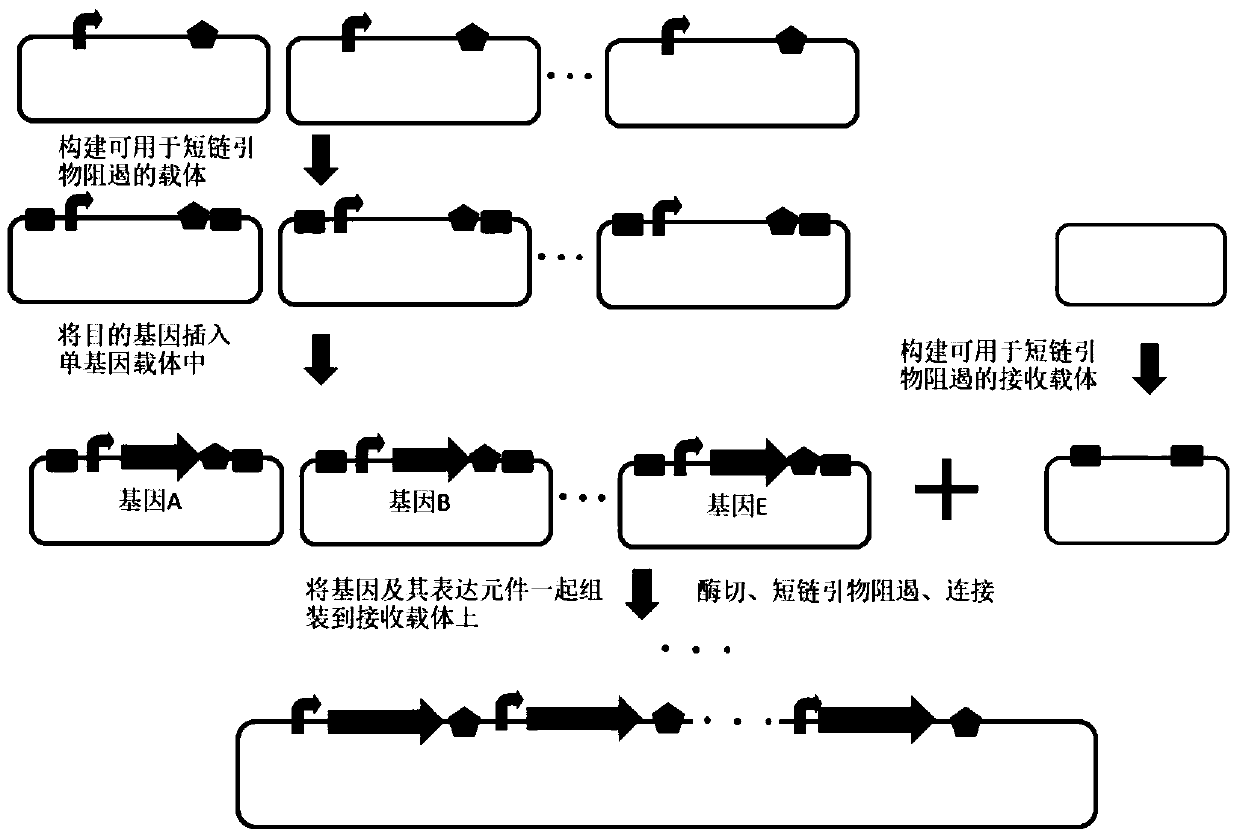
[0315] Example 2, the application of the method of short-chain primer suppression in the splicing of multiple fragments
[0316] The principle of this embodiment is attached Figure 4 shown.
[0317] 1. Preparation of 7 DNA fragments
[0318] The nucleotide sequence of DNA fragment 1 is shown as sequence 25 in the sequence table. DNA fragment 1 sequentially includes DNA fragment A1, DNA fragment 1 to be spliced and DNA fragment B1; DNA fragment A1 and short chain in DNA fragment 1 The nucleotide sequence identical or complementary to primer C is the 27th-53rd nucleotide molecule in sequence 25; the nucleotide sequence of DNA fragment 1 to be spliced is 54th-256th in sequence 25, DNA fragment B1 and The same or complementary nucleotide sequence of the short-chain primer D is the 257th-281th position in sequence 25;
[0319] The nucleotide sequence of DNA fragment 2 is shown as sequence 26 in the sequence table. DNA fragment 2 includes DNA fragment A 2, DNA fragment 2 to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com