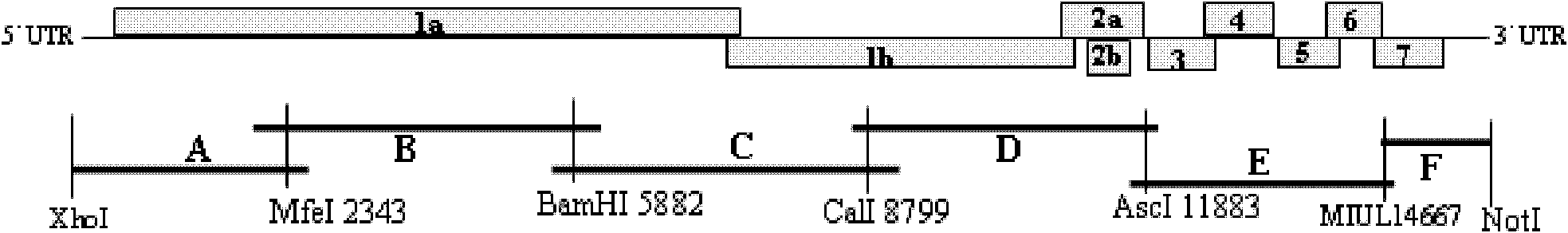
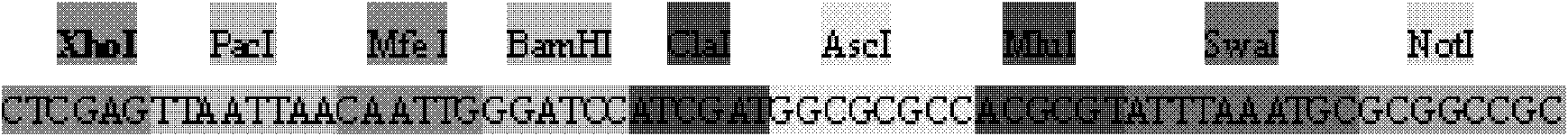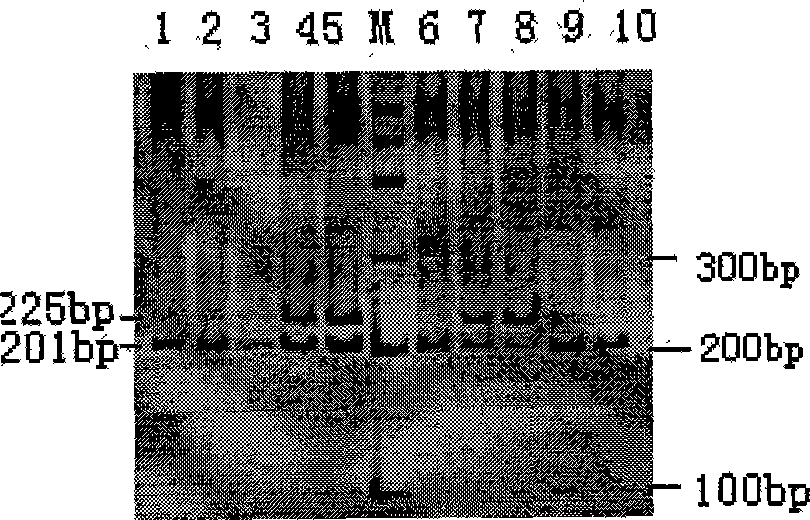Patents
Literature
77 results about "Restriction Enzyme Site" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
High-density fermentation and purification process for recombination high temperature-resistant hyperoxide dismutase
InactiveCN101275144AAvoid pollutionSimple purification processBacteriaMicroorganism based processesEscherichia coliDismutase
The present invention provides a high density fermentation and a purification process of a recombination high temperature resistance superoxide dismutase, the construction method of the invention includes: using gene coded for SOD in a thermophilic bacteria as a template, designing specific primer amplification target gene having restriction enzyme sites, after double digestion, connecting to plasmid vector pET28a after the same double digestion, constructing a recombinant plasmid, named for pSOD, transforming plasmid pSOD to competence escherichia coli BL21(DE3) by chemical transformation method, obtaining strain having high SOD yield after screening, completing the construction of SOD engineering bacteria; the fermentation process includes four steps of first order seed culture, secondary order feed culture, batch fermentation and induced expression, fermentation product SOD is finally obtained; the fermentation process realizes high level expression of SOD, the expression of the target protein is more than 60% of the bacterial protein total; SOD has excellent thermal stability and heat resistance, the expression product accounts for more than 60% of the whole proteins, and fully soluble protein, avoiding any trouble in the course of inclusion body renaturation; the purification process is simple, having high yield, lower cost, the final product SOD has high purification, high activity and strength stability.
Owner:YANGTZE DELTA REGION INST OF TSINGHUA UNIV ZHEJIANG +1
Establishment method of long mate pair library
InactiveCN102864498AQuick sortingEfficient screeningNucleotide librariesMicrobiological testing/measurement3-deoxyriboseDeoxyribose
The invention relates to an establishment method of a long mate pair library. The establishment method comprises the steps as follows: 1) randomly interrupting DNA (deoxyribose nucleic acid), and completing and repairing the gaps at the tail end; 2) preparing A-Fragment; 3) carrying out cohesive end connection to A-Fragment and LMP Adaptor; 4) digesting the LMP-Adaptor-Fragment with at least two restriction enzymes which can distinguish four basic groups; 5) cyclizing the fragments in the obtained enzyme digesting library again; and 6) amplifying the purified ring molecule through primer of the LMP Adaptor; and recovering the amplified products, thus finishing the establishment of LMP (long mate pair) library. According to the library establishment method provided by the invention, only two 63bp oligonucleotides sequences are needed to be synthetized, and simple experiments such as molecular biology connection and amplification are carried out, and the two ends of the obtained LMP library have the sequencing primer sequences which can be directly applied to next generation sequencing; and moreover, the fragment subjected to secondary cyclizing has the known restriction enzyme site, and the sequences of two ends of the to-be-tested fragments can be quickly sorted, the sequence testing data can be effectively sieved, and the mosaic sequence can be removed.
Owner:TIANJIN INST OF IND BIOTECH CHINESE ACADEMY OF SCI
Engineering bacterium capable of producing anthracene ring antibiotics and application of the same
The invention discloses an engineering bacterium through anthracene nucleus antibiotic and appliance, which is characterized by the following: inserting or replacing restriction enzymes site of dnmV gene nucleic acid order of sky blue rufus streptomycete with antibiotic against property gene and surface cerubidin synthetic gene; blocking-up engineering bacterium of dnmV gene; or transforming expressing carrier of anthracene nucleus antibiotic synthetic gene to lead streptomycin (S.lividans)TK24; extracting expressing plasmid from S.lividans TK24; leading-in sky blue rufus streptomycete gene only with antibiotic against property gene to block dnmV gene; blocking-up abrupt change bacterium. This invention can produce cerubidin and / or analogue, such as anthracene nucleus antibiotic.
Owner:ZHEJIANG HISUN PHARMA CO LTD +1
Vaccine strains of infectious clones of porcine reproductive and respiratory syndrome virus (PRRSV) and application thereof
ActiveCN101984061AEasy to solveEasy to controlViral antigen ingredientsMicrobiological testing/measurementHighly pathogenicAttenuated vaccine
The invention discloses an artificially cloned attenuated vaccine strains. The vaccine strains are strains cloned from attenuated vaccine strains HuN4-F112 of highly pathogenic porcine reproductive and respiratory syndrome virus (PRRSV), and marked restriction enzyme sites are introduced in the structural protein gene sequences of the strains. The invention also discloses a recombinant vector which comprises a full-length gene cDNA sequence of the attenuated vaccine strains HuN4-F112 of the highly pathogenic PRRSV. The 5'-end of the full-length gene cDNA sequence is additionally provided witha transcription promoter and the full-length gene cDNA sequence is internally introduced with the marked restriction enzyme sites. The artificially cloned attenuated vaccine strains of the invention can not only provide completely safe immune protection for resistance of an immune pig to the highly pathogenic PRRSV, but also effectively distinguish an immune pig of the PRRSV from a naturally infectious pig of the PRRSV, thus being beneficial to preventing and controlling the highly pathogenic PRRSV.
Owner:SHANGHAI VETERINARY RES INST CHINESE ACAD OF AGRI SCI
Inducible expression, purification and activity identification method of restructured lunasin polypeptide in pichia pastoris
InactiveCN105647960ALow costIncrease productionDiagnosticsMicrobiological testing/measurementHigh resistancePurification methods
The invention discloses an inducible expression, purification and activity identification method of restructured lunasin polypeptide in pichia pastoris. The inducible expression and purification method includes 1) establishing the restructured expression plasmid: designing Lunasin sequence with EcoR I and Not I restriction enzyme sites, and designing specificity ampflification primer for PCR (polymerase chain reaction) amplification to obtain target gene segments; 2) screening high-resistance restructured gene engineering strains; 3) fermenting the restructured lunansin polypeptide; 4) separating and purifying the restructured lunansin polypeptide: obtaining the restructured lunasin polypeptide with purity at 93%. The pichia pastoris is firstly used to prepare lunasin polypeptide, and batch production of the lunasin polypeptide can be realized through the gene engineering method. The lunasin polypeptide expressed by the pichia pastoris is low in cost, and high in production, and separation and purification steps are simple.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
General type gene targeting vector
ActiveCN104651402AReduce expressionExpression capacityVector-based foreign material introductionFluorescenceCloning Site
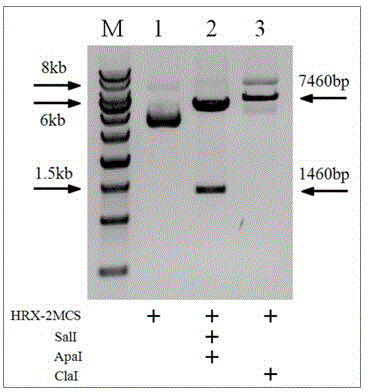
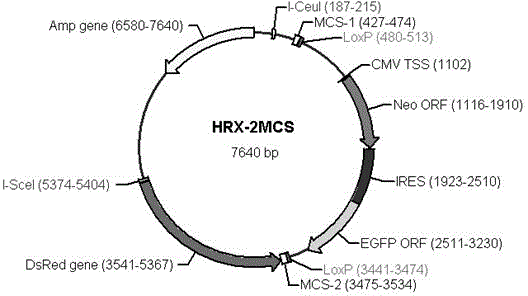
The invention discloses a general type gene targeting vector, and belongs to the field of bioengineering. The general type gene targeting vector uses neomycin phosphotransferase and EGFP as positive selective markers and DsRed as a negative selective marker; two sides of a positive selective marker gene expression frame respectively contain LoxP sites which are arranged in a same direction; and the LoxP sites respectively contain multiple cloning sites which include rare restriction enzyme sites nearby, so that the convenience for the cloning of a gene targeting arm is achieved. The general type gene targeting vector contains two homing restriction enzyme sites, and the two homing restriction enzyme sites are respectively I-CeuI and I-SceI and can be used as general endonuclease sites for linearizing the vector. The general type gene targeting vector provided by the invention not only has the functions of single drug and double fluorescent screening, but also provides a new material for an animal safety transgenic technology because the contained LoxP sites are capable of mediating the high-efficiency deletion reaction of the selective markers, and has significant application prospects.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
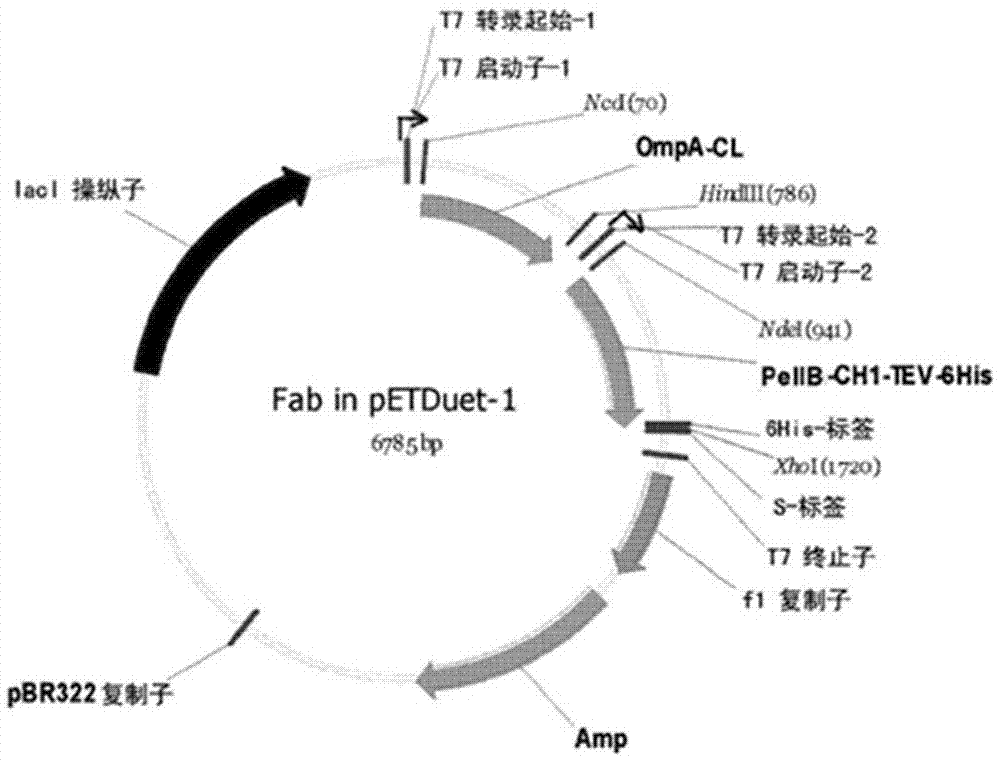
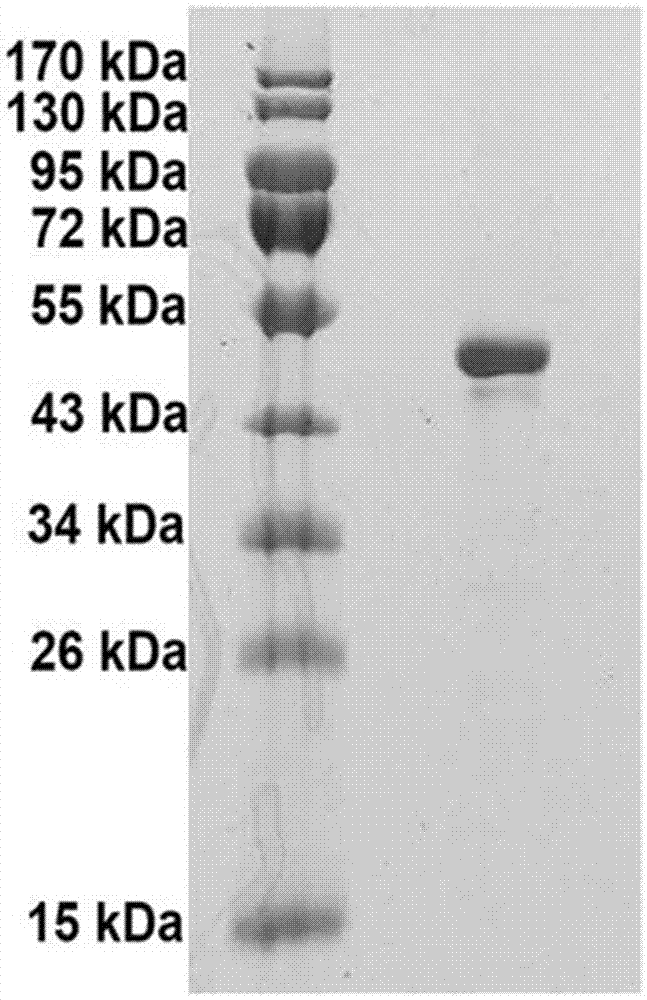
Fab fragment of fully-human recombinant anti-CD40L monoclonal antibody and preparation method thereof
ActiveCN106967171AHigh purityFast preparationImmunoglobulins against cell receptors/antigens/surface-determinantsPeptide preparation methodsDiseaseSequence signal
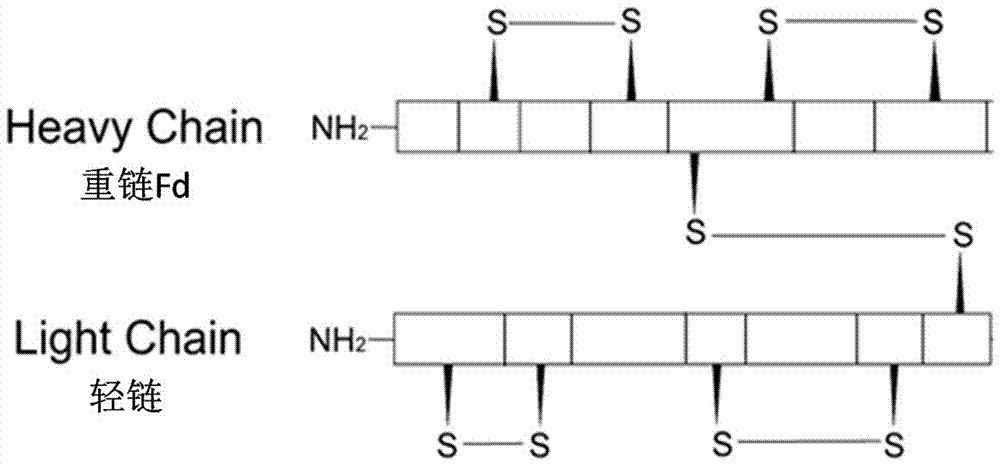
The invention specifically relates to a Fab fragment of a fully-human recombinant anti-CD40L monoclonal antibody and a preparation method thereof, belonging to the field of genetic engineering technology. The Fab fragment comprises a heavy chain Fd and a light chain. During expression and preparation of the Fab fragment, a signal peptide PelB is added to the N terminal of the heavy chain Fd, and the C terminal of the heavy chain Fd is connected with a protease TEV restriction enzyme site and a histidine fusion tag; a signal peptide OmpA is added to the N terminal of the light chain; then a recombinant expression vector is constructed after connection with an Escherichia coli expression vector pETDuet-1 with double promoters; and the Fab fragment is further expressed and prepared. According to the invention, the Fab fragment is optimized and a host cell, the expression vector, expression conditions and purification conditions are reconstructed and optimized, so the Fab fragment is rapidly prepared, has the technical advantages of high purity, low cost, stable effect and the like and has good application value in prevention and control of related diseases.
Owner:ZHENGZHOU UNIV
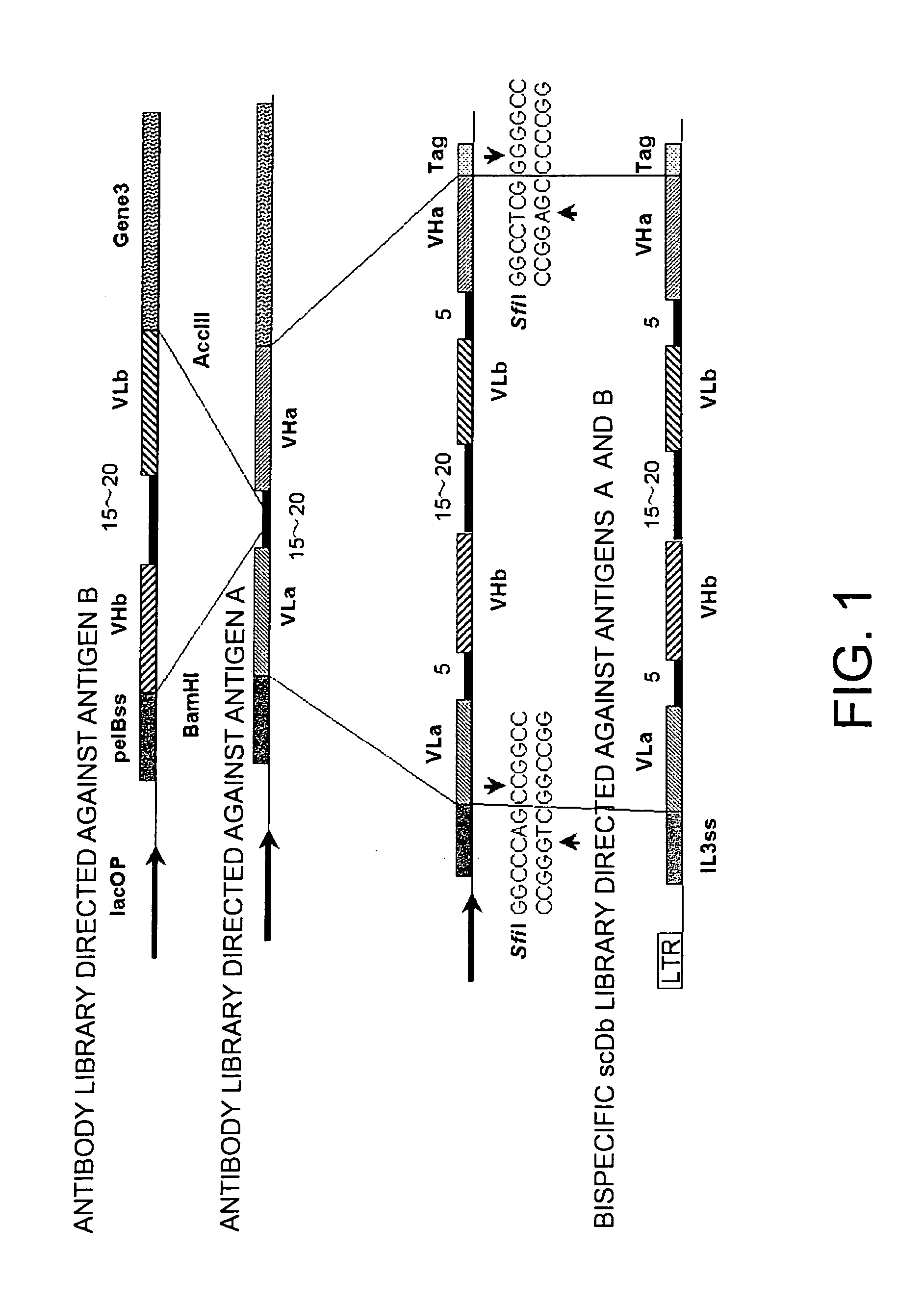
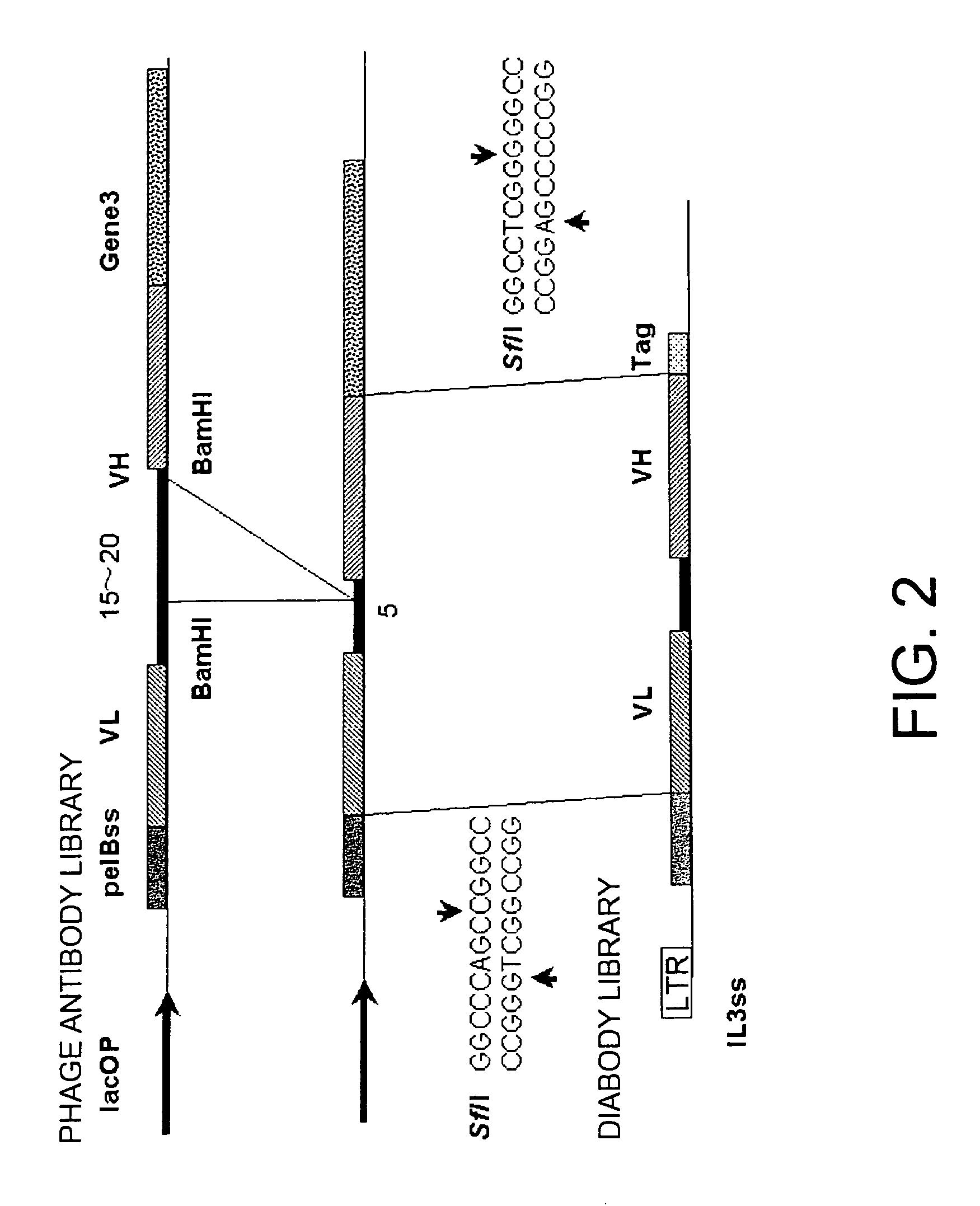
Methods for constructing scDb libraries
Owner:CHUGAI PHARMA CO LTD
Vector for efficiently secreting and expressing heterogenous protein, and its application
InactiveCN104342445AImprove build efficiencyShorten the timeBacteria peptidesEnzymesSequence signalSolubility
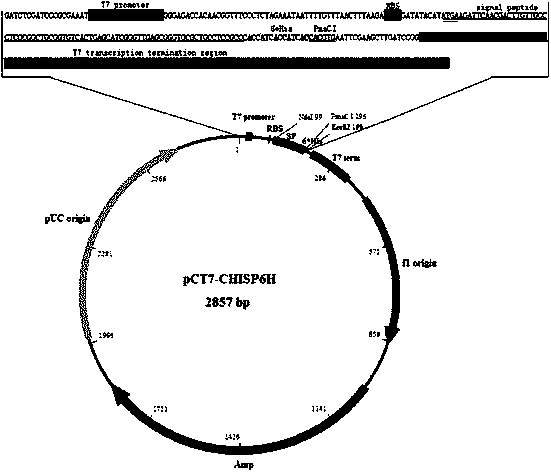
The invention relates to an expression vector in the gene engineering field, and concretely relates to a vector for efficiently secreting and expressing a heterologous protein, and its application. An encoding signal peptide for efficiently secreting and expressing the heterologous protein is represented by a base sequence in SEQID NO.1. The vector for efficiently secreting and expressing the heterologous protein contains a specific nucleotide sequence, the sequence is the nucleotide sequence of a promoter and the encoding signal peptide; structure diagrams comprise a promoter sequence, a nucleotide sequence encoding the signal peptide, a 6*His tag sequence and a PmaCI restriction enzyme site; and the promoter is a T7 promoter, and the encoding signal peptide is a chitosan enzyme signal peptide of microbacterium (Microbacterium sp.OU01). The vector can efficiently secreting and expressing exogenous gene and also can effectively improve the water solubility of expression products, and a recombinant protein with biological activity can be obtained by using affinity chromatography one-step purification.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Mammalian retrotransposable elements
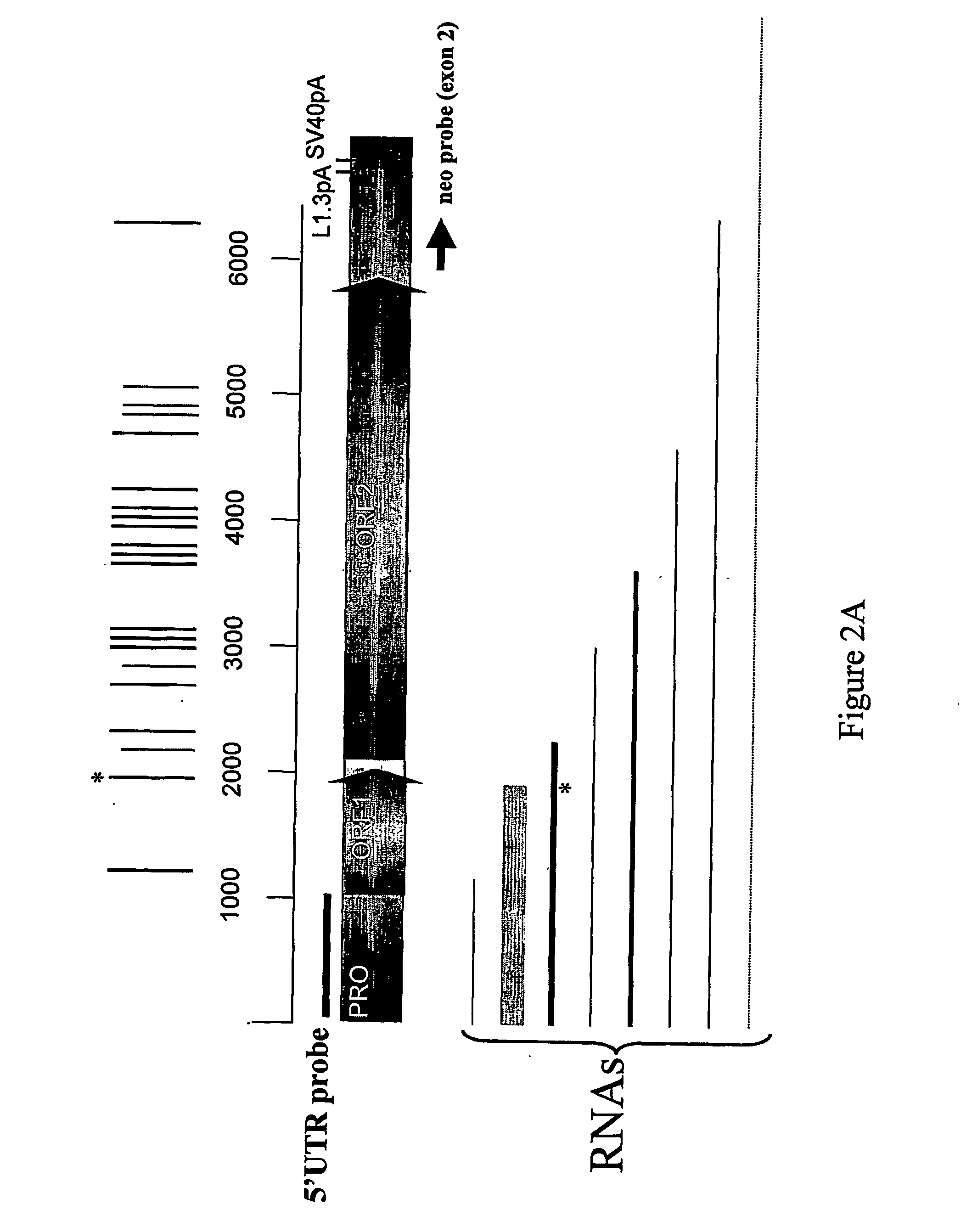
InactiveUS20070037759A1High efficiency of retrotranspositionIncrease gene expressionGenetic material ingredientsStable introduction of DNAPoly-A RNAMammal
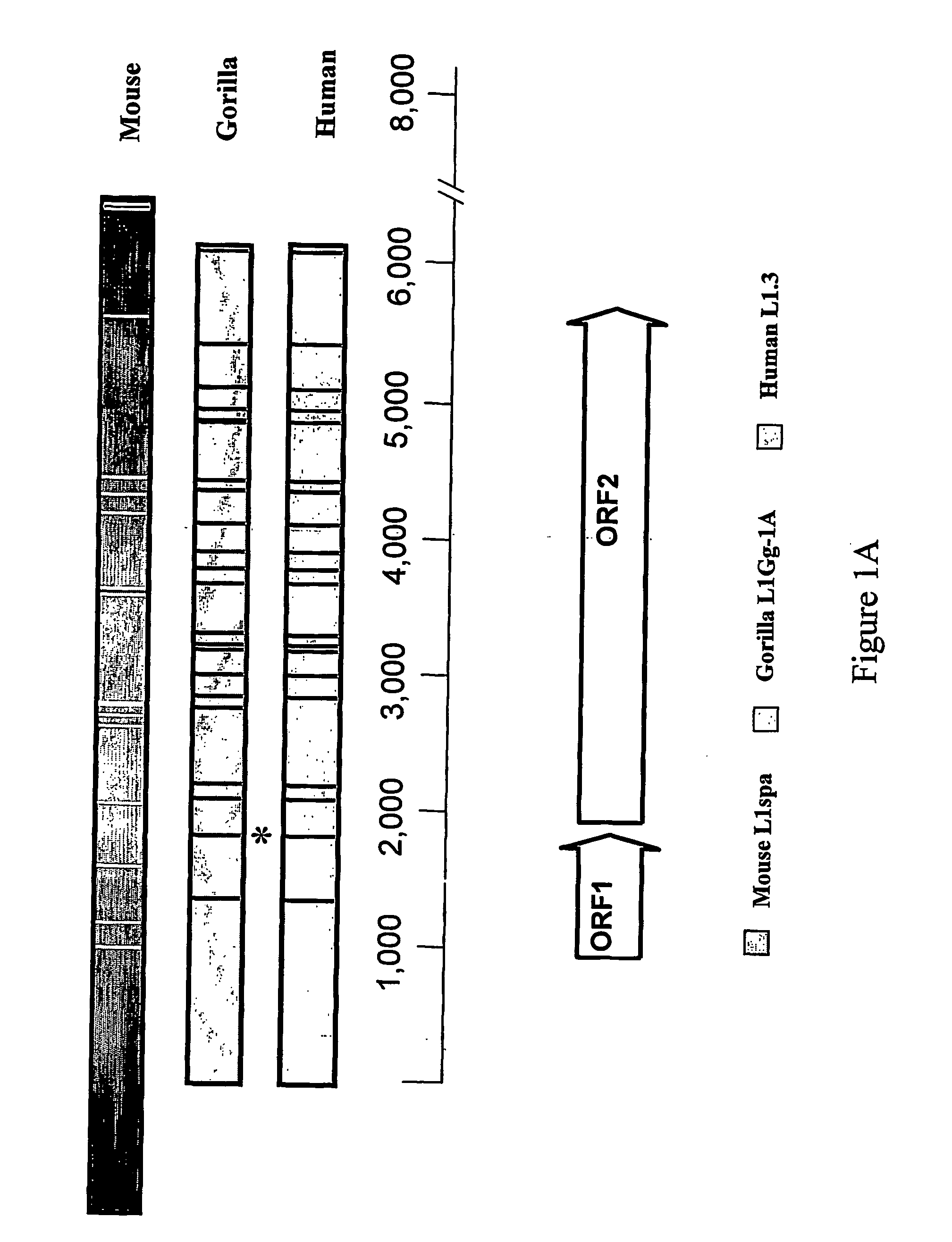
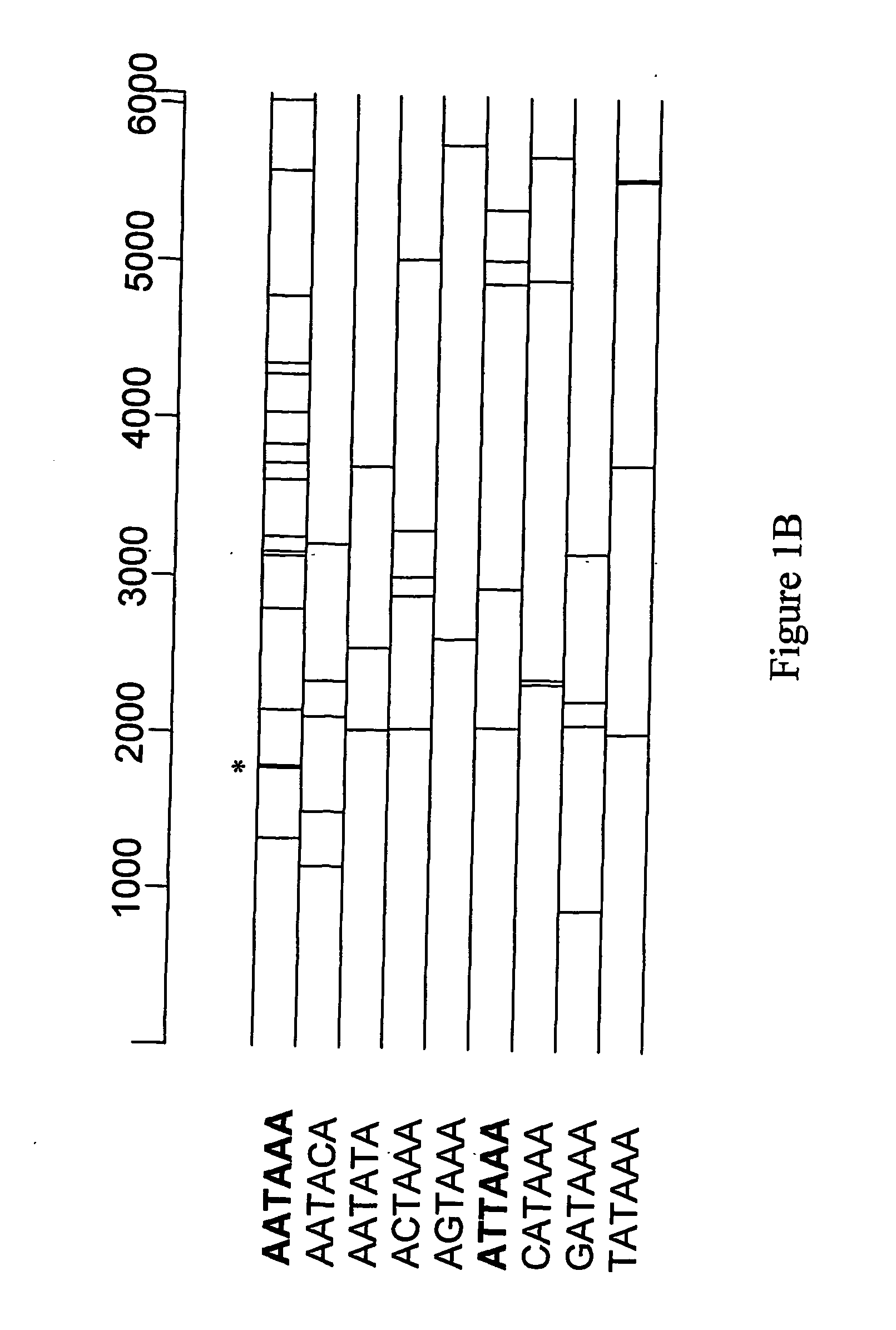
The invention relates to a modified retrotransposable element wherein one or more polyadenylation sites have been removed from within the genes of the retrotransposable element, and methods of use thereof. Sequences of optimized retrotransposable element are provided that have polyadenylation sites removed, and may be further modified to optimize codon usage for expression in mammals and to add restriction enzyme sites for ease of use.
Owner:TULANE EDUCATIONAL FUND
Expression vector, host, fused protein, process for producing fused protein and process for producing protein
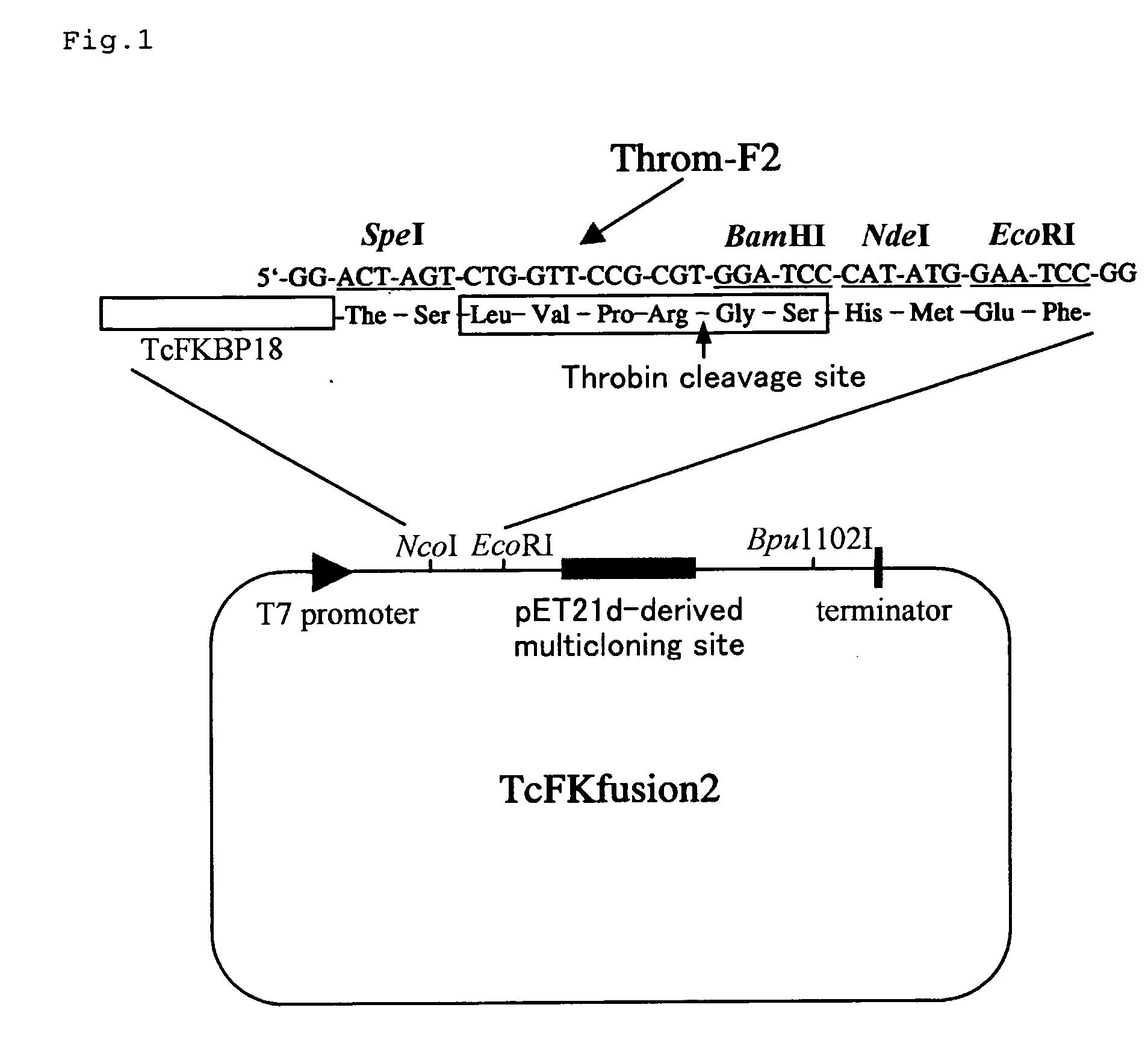
InactiveUS20050130259A1Inhibition formationBacteriaSugar derivativesCoding regionRestriction Enzyme Site
It is an object of the present invention to provide an expression vector, a host, a fused protein, a protein, a process for producing a fused protein, and a process for producing a protein, which can prevent formation of an unactive abnormal protein at production of a recombinant protein, and can produce a desired protein as a natural type, that is, a soluble type at a large amount and effectively. That is, the present invention is an expression vector, which comprises: (a) a first coding region encoding a polypeptide having molecular chaperone activity, and (b) a region having at least one restriction enzyme site in which a second coding region encoding a protein can be inserted. In the expression vector of the present invention, the first coding region is operatively linked to a promoter, and the restriction enzyme sites are in the same reading frame as the first coding region, and are downstream of the first coding region, or the restriction enzyme sites are disposed so that the inserted second coding region is operatively linked to a promoter, and the first coding region is in the same reading frame as the second coding region, and is downstream of the second coding region.
Owner:SEKISUI CHEM CO LTD
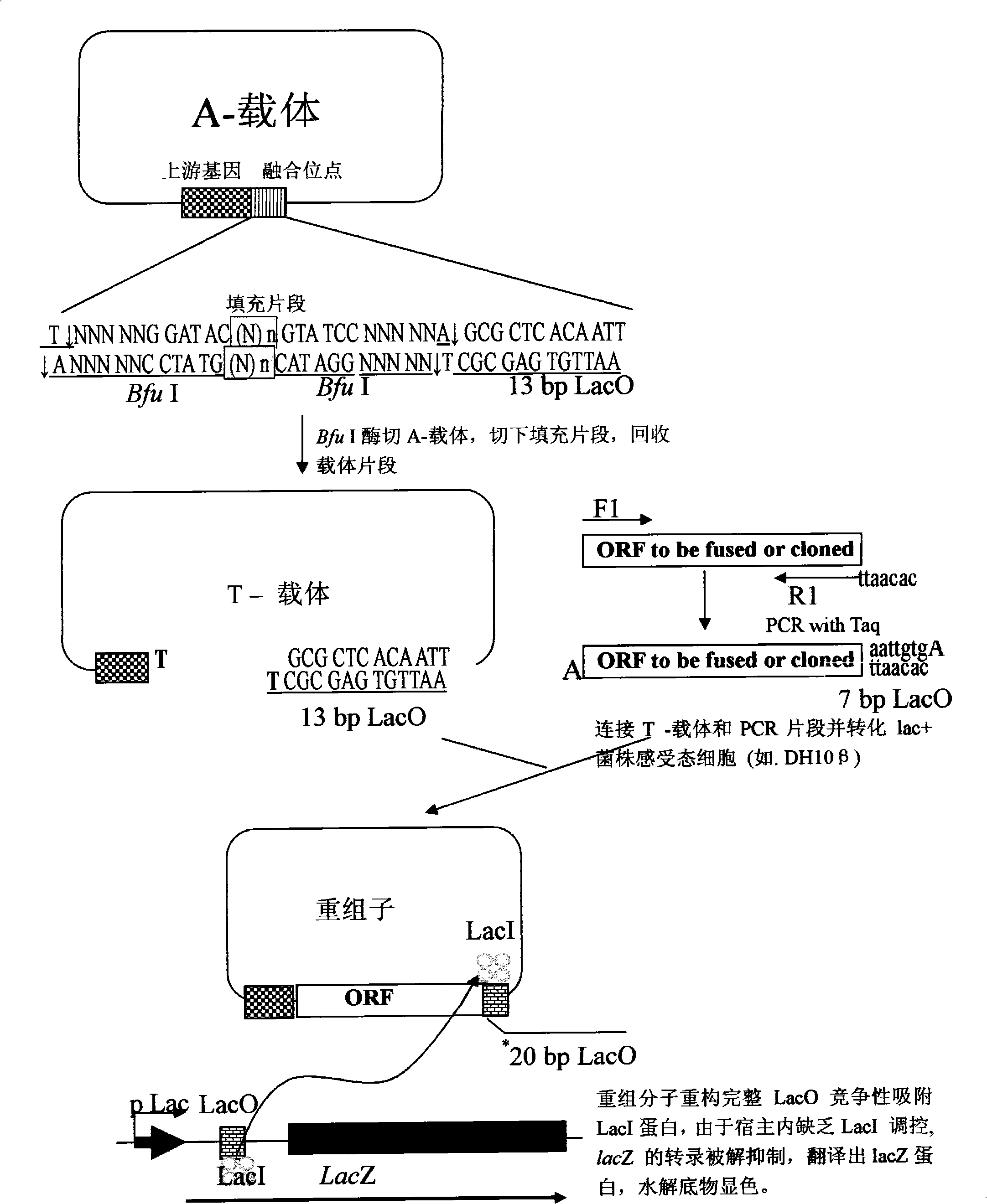
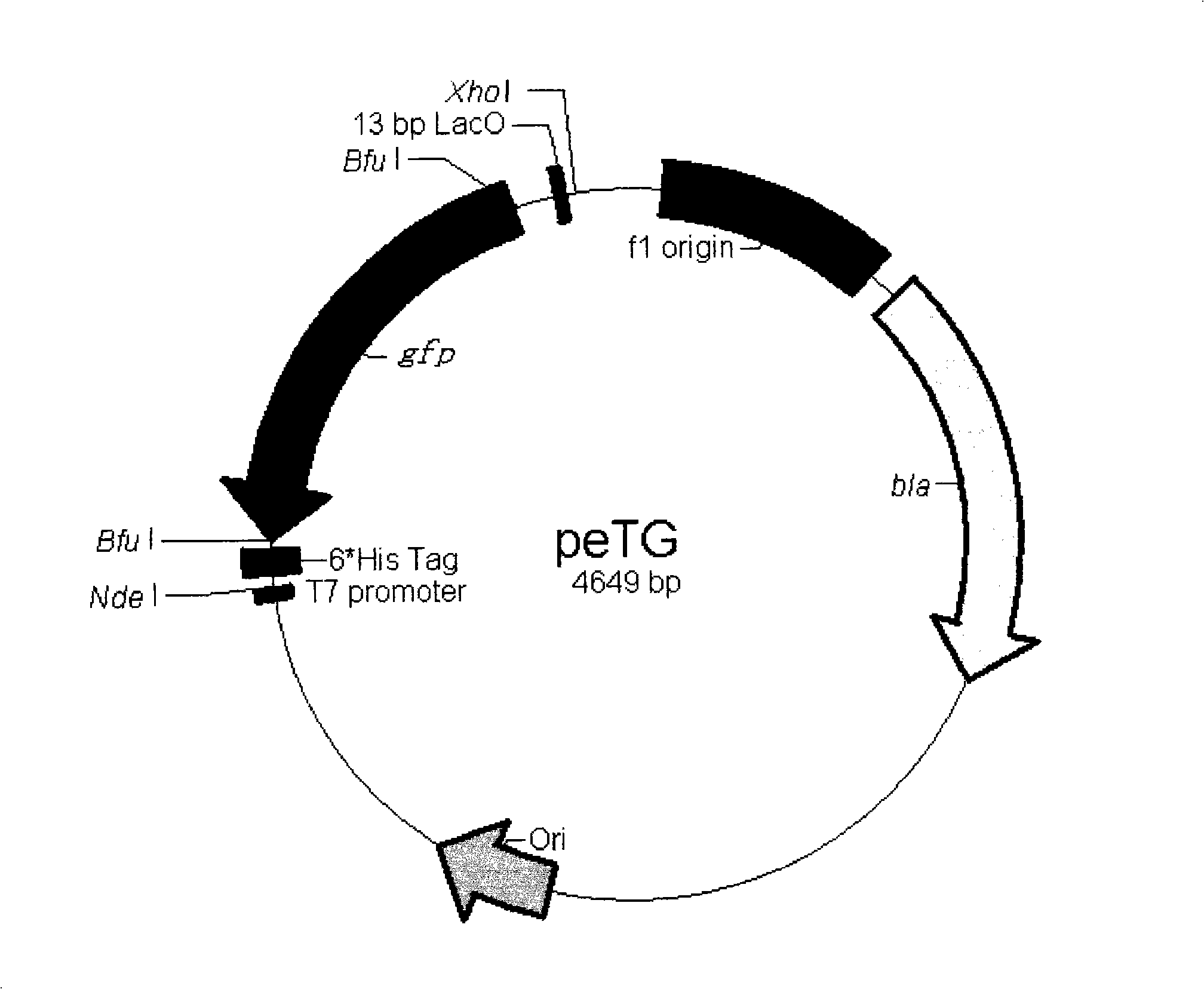
High throughput directional T carrier cloning process
InactiveCN101307321ASimple designFlexibilityVector-based foreign material introductionHigh fluxOperator gene
The invention provides a method for cloning high-flux directed T vector. The method is as follows: an appropriate IIS type restriction enzyme site and lactose operator gene sequence (LacO) are selected; through reconstructing integrate lactose operator gene sequence (LacO) between vector mutagenesis cleavage joint transformation and cloning fragment, directed cloning and non-identification cloning are realized; moreover, fusion and non-fusion T vector construction is realized. The method has simple primer design, high cloning efficiency, less time consumption and low cost; moreover, the obtained recon does not need subsequent identification and is compatible with the prior cloning method.
Owner:HUBEI HUAGUOSHAN IND
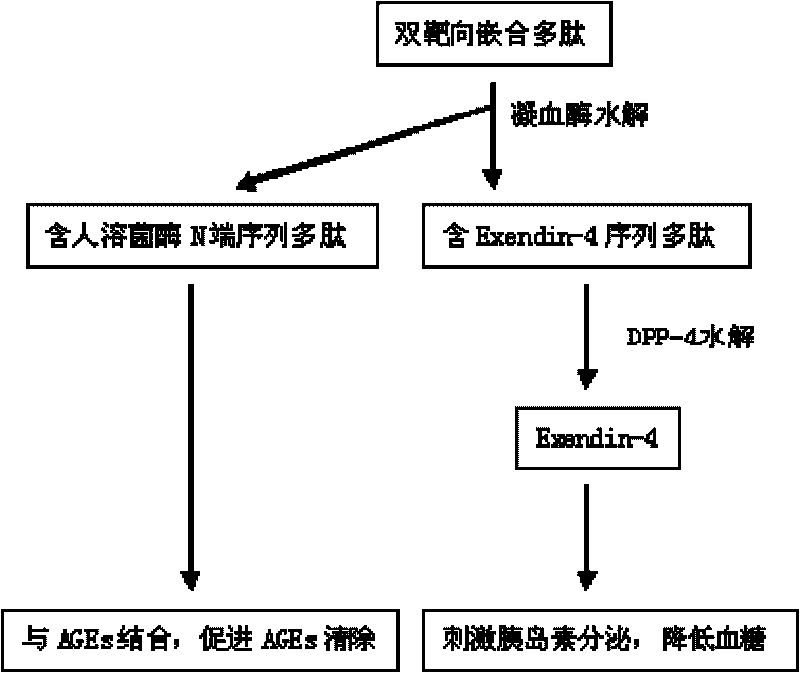
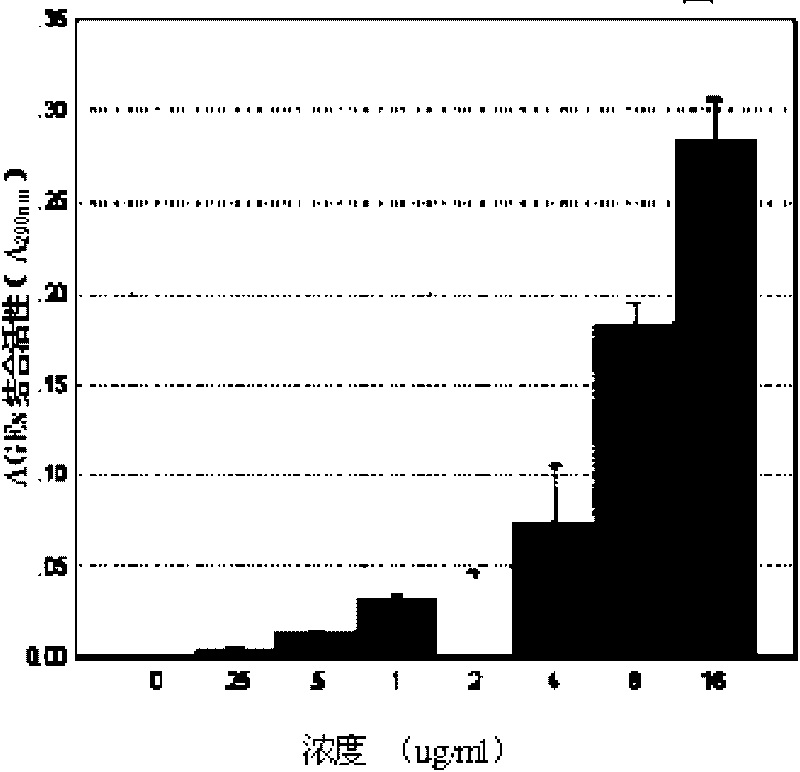
Chimeric polypeptide with dual-targeting function and applications thereof
InactiveCN101698681AHigh activitySignificantly anti-AGEsPeptide/protein ingredientsMetabolism disorderDipeptidyl peptidaseNucleotide
The invention provides a chimeric polypeptide with dual-targeting function, and a preparation method and applications thereof. The chimeric polypeptide of the invention comprises three sequences with the following characteristics: sequence 1 is an amino acid sequence comprising the first 20-80 human lysozyme proteins counting from end N; sequence 2 is an amino acid sequence of Exendin-4 proteins; and sequence 3 is a connecting sequence of the sequence 1 and the sequence 2 and comprises a thrombin restriction enzyme site and a dipeptidyl peptidase restriction enzyme site. The chimeric polypeptide can be synthesized by using a polypeptide synthesizing technology, or can be obtained by constructing an expression carrier comprising the polypeptide nucleotide sequence specified by the code, expressing in a genetic engineering stain and finally purifying. The chimeric polypeptide of the invention has obvious dual-targeting activity for resisting AGEs and reducing blood sugar, and can be used for preparing medicines for resisting AGEs and / or lowering high blood sugar.
Owner:JINAN UNIVERSITY
A segment of sequence and a restriction enzyme site for differentiating canine distemper viral vaccine strain and wild strain
InactiveCN101376909AEasy to controlIdentification method is simpleMicrobiological testing/measurementRestriction Enzyme Cut SiteElectrophoresis
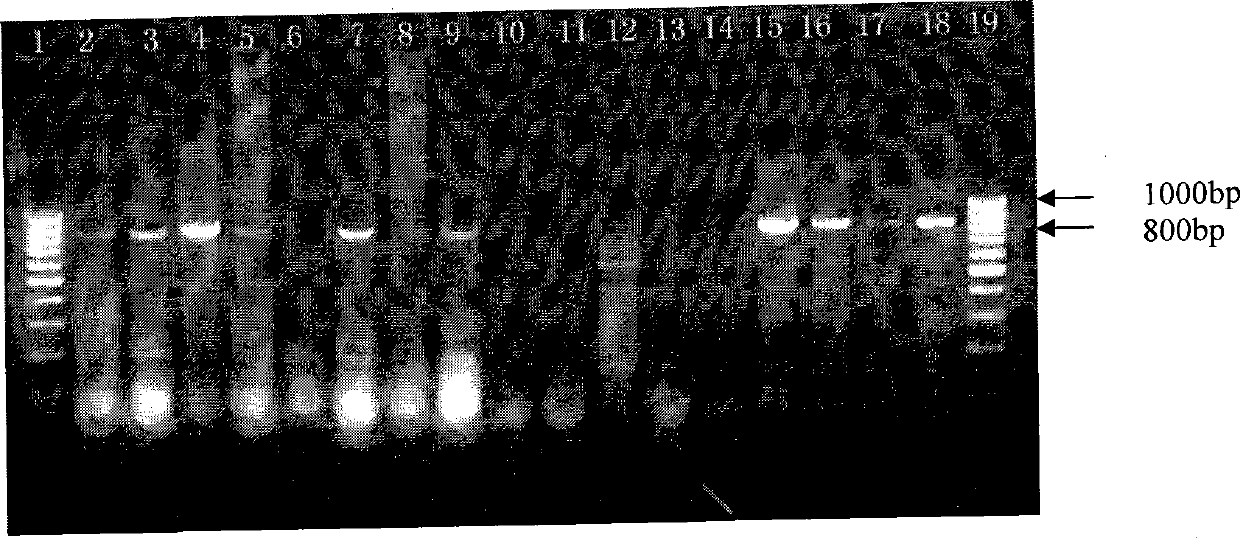
The invention relates to a method used to distinguish a sequence and a restriction enzyme cutting site of a canine distemper virus vaccine strain and a field strain and integrate two molecular biology test methods, namely RT-PCR and RFLP; a CDV NP gene is selected; a special primer is designed to amplify 829bp, and the segment is treated by BamHI RFLP; through agarose gel electrophoresis judgment, the canine distemper virus tested in the animal body is identified to belong to an inoculated vaccine strain or be injected by field virus from the number of segment; two segments 675bp and 153bp obtained from the electrophoresis are vaccine strains, and three segments 455bp, 220bp and 153bp are field strains. The method effectively distinguishes the canine distemper virus vaccine strain and the field strain, and provides veterinary technical guarantee for the cultivation of fur animals.
Owner:INST OF SPECIAL ANIMAL & PLANT SCI OF CAAS
Reverse restriction fragment length polymorphism assay and uses thereof
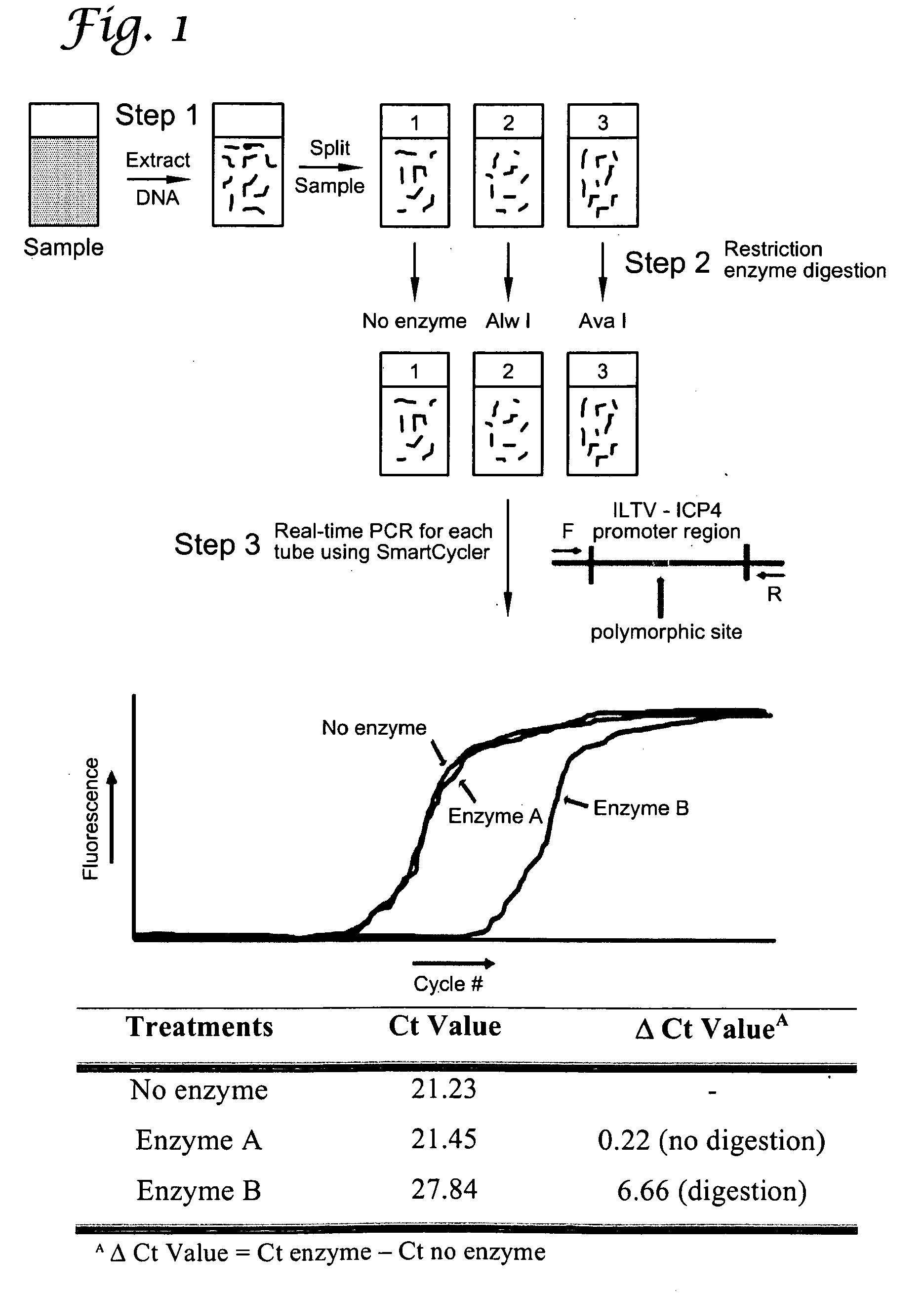
InactiveUS20100196881A1Microbiological testing/measurementRestriction enzyme digestionTerminal restriction fragment length polymorphism
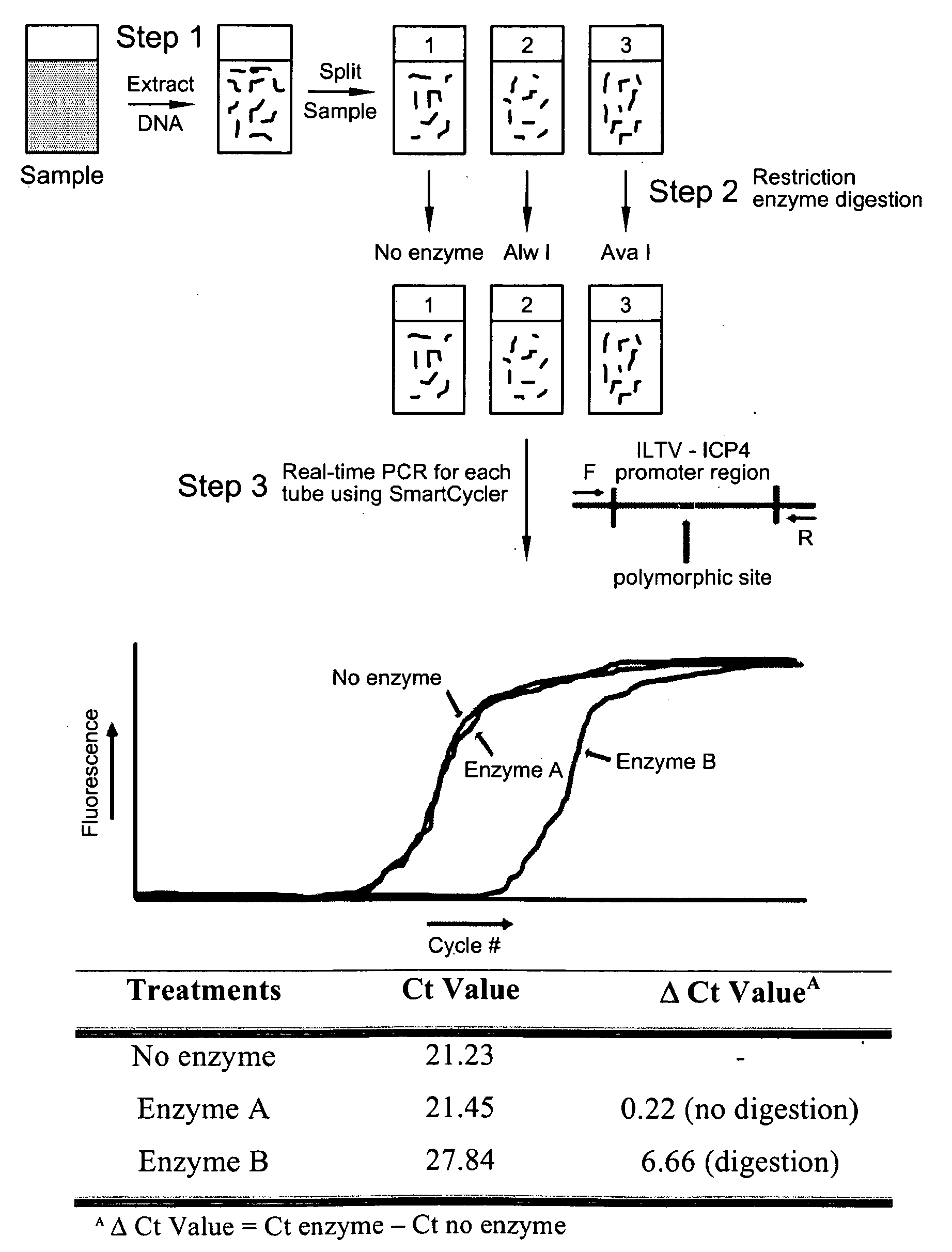
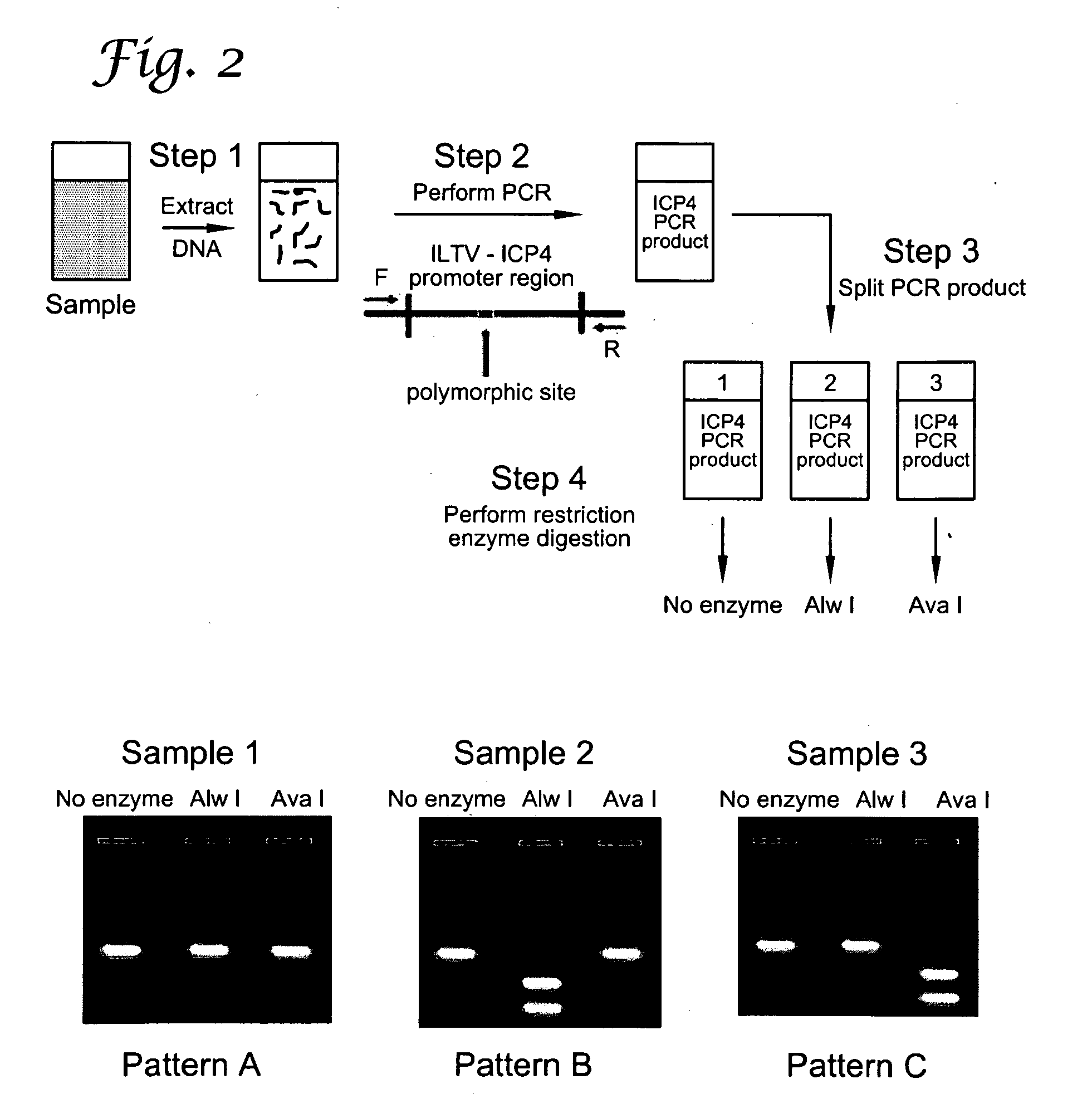
The present invention presents a Reverse Restriction Fragment Length Polymorphism (RRFLP) method for the detection of the presence of an informative restriction enzyme site in a nucleotide sequence. The method includes digesting a sample with the informative restriction enzyme; performing polymerase chain reaction (PCR) on the digested sample with an oligonucleotide primer pair that flanks the informative restriction enzyme site; determining the Ct value of the sample; comparing the Ct value of the sample to the Ct value from a control sample; and calculating a ΔCt value, wherein a ΔCt value is the Ct value of the sample minus the Ct value of a control; and wherein a ΔCt value ≧+1 indicates that the informative restriction enzyme sites is present in the nucleotide sequence. The present invention includes the application of the RRFLP method for detection of the infectious laryngotracheitis virus (ILTV).
Owner:UNIV OF GEORGIA RES FOUND INC
Chromatin conformation capturing method
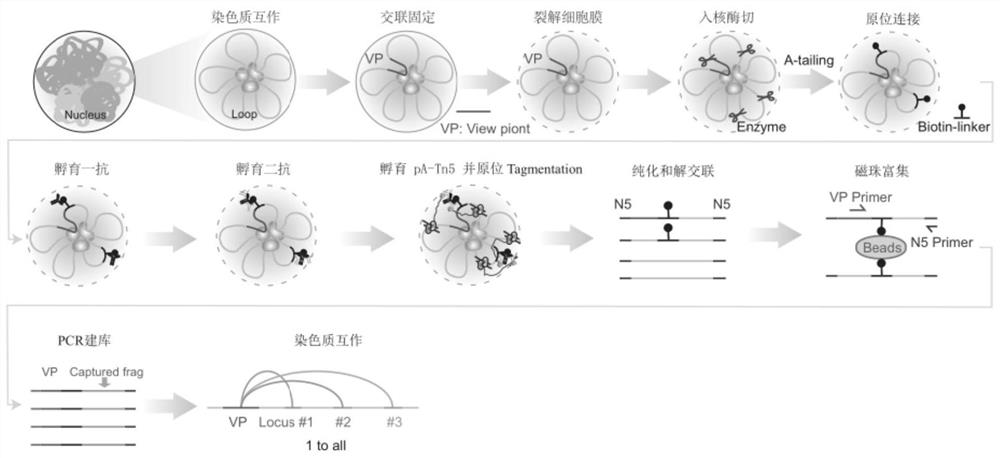
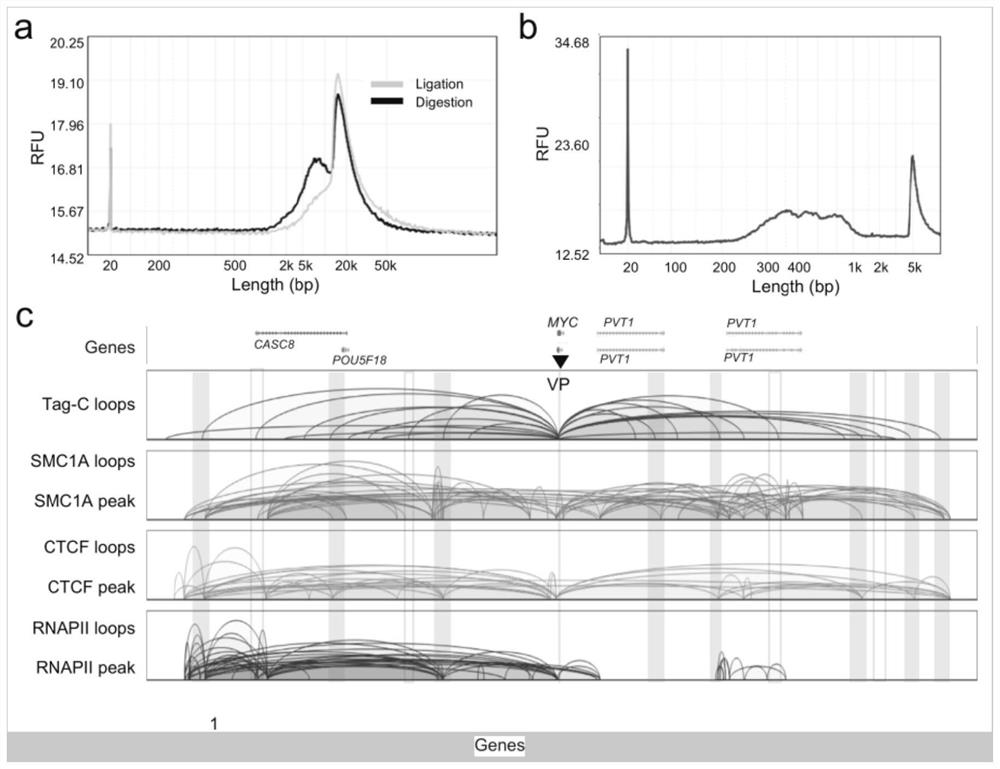
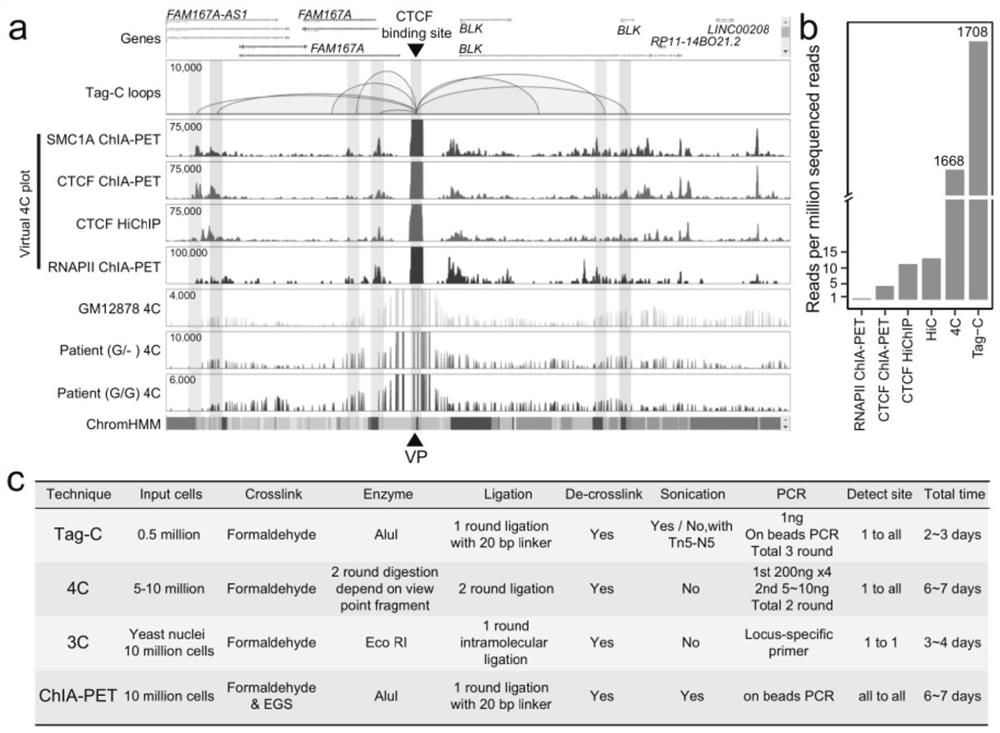
ActiveCN113466444ASimplify build stepsImprove featuresBiological testingLibrary creationEnzyme digestionBiotin
The invention relates to the field of biology, and particularly discloses a chromatin conformation capturing method. The method comprises the steps of carrying out an in-situ proximity ligation reaction in a cell and labeling a ligation position with biotin; identifying an adjacent ligation position by using biotin antibody and secondary antibody specificity, binding Tn5 in pA-Tn5 to DNA near the adjacent ligation position in a targeted manner through binding of pA and antibody, breaking the DNA near the adjacent ligation position by Tn5 enzyme digestion, and simultaneously accessing an N5 linker sequence; and after the DNA is purified, enriching DNA with biotin by using magnetic beads as a template, performing PCR amplification on the template DNA by using a specific primer of an observation site and a P5 primer to construct a sequencing library, sequencing and analyzing data. According to the method, the dependency of the observation site on the restriction enzyme site is greatly reduced, the chromatin three-dimensional conformation in which the observation site participates is detected with higher resolution, the number of required cells is greatly reduced, and the method can be applied to clinical samples.
Owner:SUN YAT SEN UNIV
Method for quickly constructing recombinant plasmid
InactiveCN106497958ASimplify build stepsWon't interfereVector-based foreign material introductionBiotechnologyEnzyme digestion
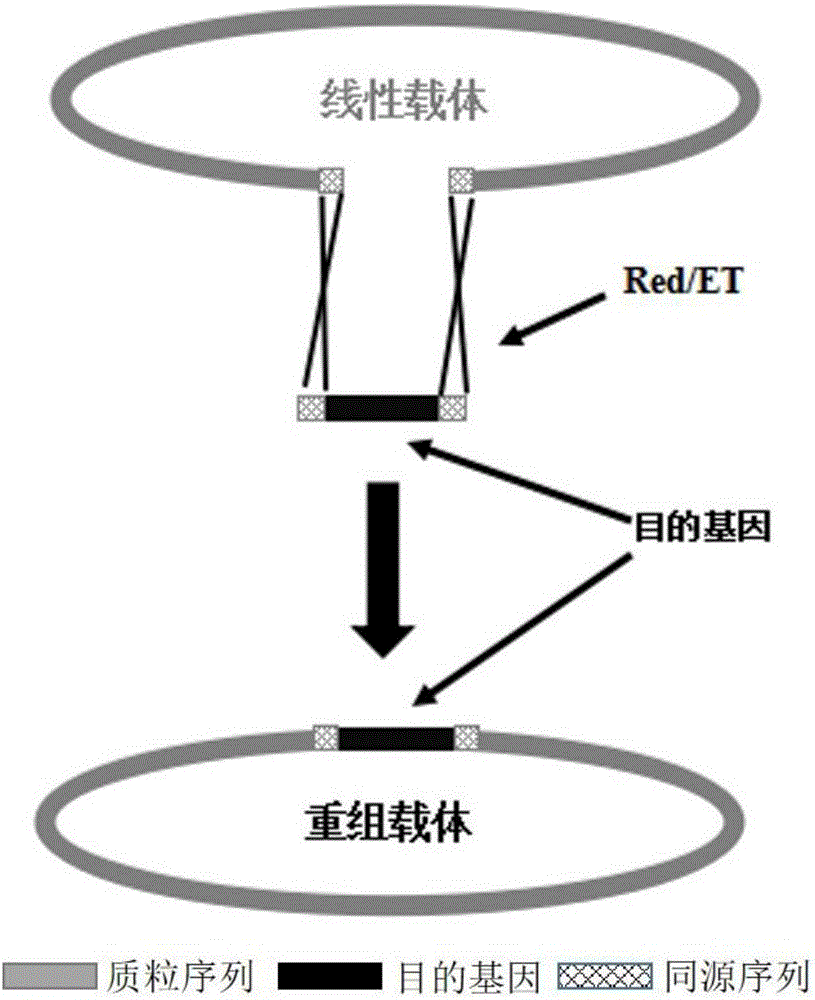
The invention belongs to the technical field of genetic engineering and discloses a method for quickly constructing a recombinant plasmid; the method comprises the steps of designing homologous recombinant primers, preparing E. coli competent cells carrying pKD46 plasmid, and co-converting an exogenous DNA fragment having sections homologous to the plasmid. An E. coli strain with Red recombinant system is constructed by using pKD46 plasmid to stably express Red recombinase under the induction of L-arabinose, the recombinase is capable of promoting the sections homologous to the plasmid and carried to two ends of the exogenous DNA fragment to recombine with a linear plasmid, it is achieved for the first time that an exogenous DNA fragment is inserted into a single limited restriction enzymes site of a support by means of RED recombination technology, it is possible to eliminate pKD46 plasmid by cultivation at 37 DEG C, no disturbance is caused to subsequent experiments, the construction steps of the recombinant plasmid are greatly simplified, and construction time is shortened. The produced recombinant plasmid is verified via PCR (polymerase chain reaction) and enzyme digestion to exhibit high positive cloning efficiency, and the method may act as a new means to construct a recombinant plasmid.
Owner:WUHAN INST OF BIOENG
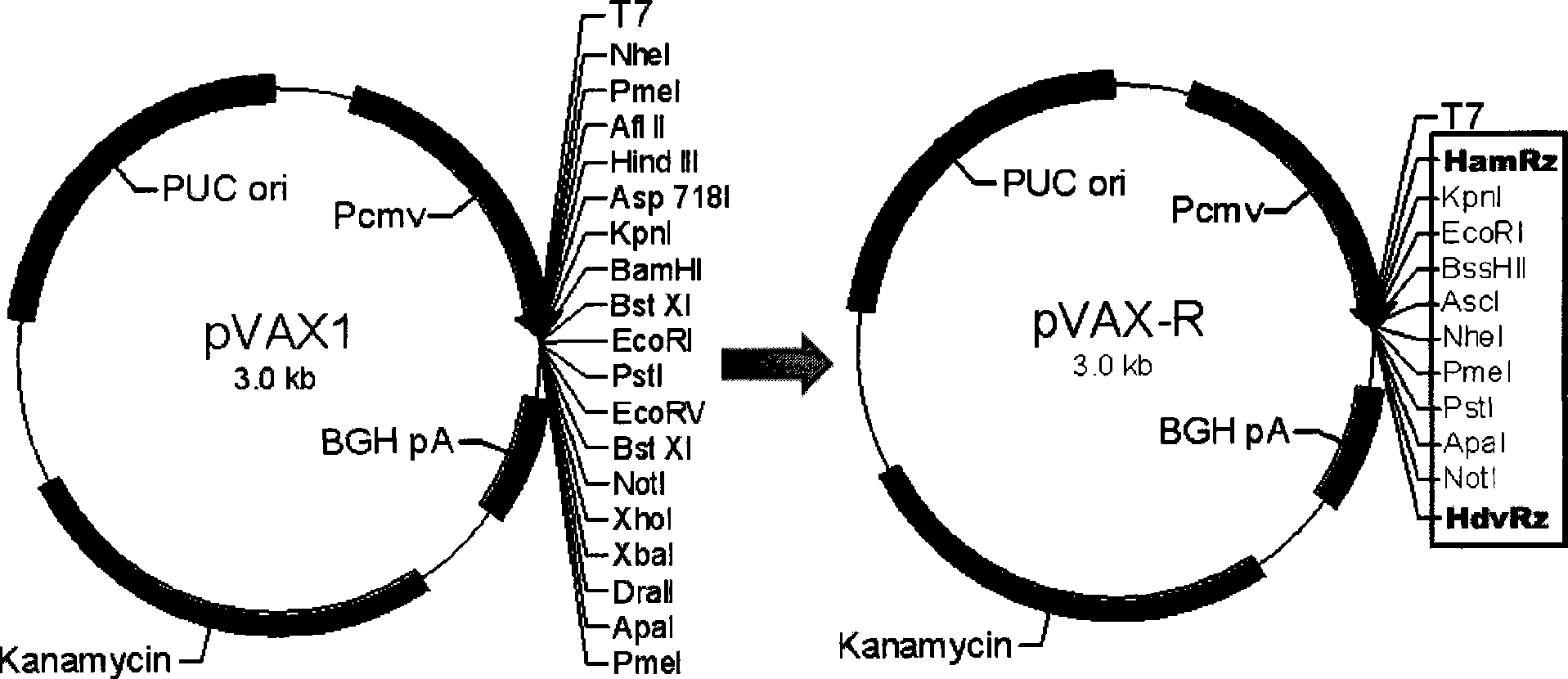
Rabies viruses CTN strain complete genome infectious cDNA cloning, and preparation and use thereof
InactiveCN101475943APreserve weak toxicitySafeMicroorganism based processesAntiviralsCdna cloningViral vector
The present invention provides a rabies virus CTN strain whole genome cDNA infectious clone, as well as a method of using reverse genetics technology to save the active rabies virus. The present inventive method includes: using the single restriction enzyme sites in the CTN strain of rabies virus full-length genome to divide the full-length virus into four segments to amplify, simultaneously using the green fluorescent protein gene to substitute 423bp base at the pseudo-gene point, cloning in the pVAX-R expression vector, to construct a recombinant full-length plasmid of the virus genome; performing PCR amplification of nucleoprotein, phosphoprotein, glycoprotein and transcription large protein gene sequence of the CTN strain of the rabies virus, and cloning the amplified fragment into a vector pVAX1, to constitute four auxiliary plasmids in the reverse heredity system; and then using the five plasmids to jointly transfect cells, to rescue the rabies virus. The present invention successfully saves the rabies virus, and has a great significance in the study of the pathogenic mechanism of rabies virus, development of new vaccines, viral vectors and the like areas.
Owner:中国疾病预防控制中心病毒病预防控制所
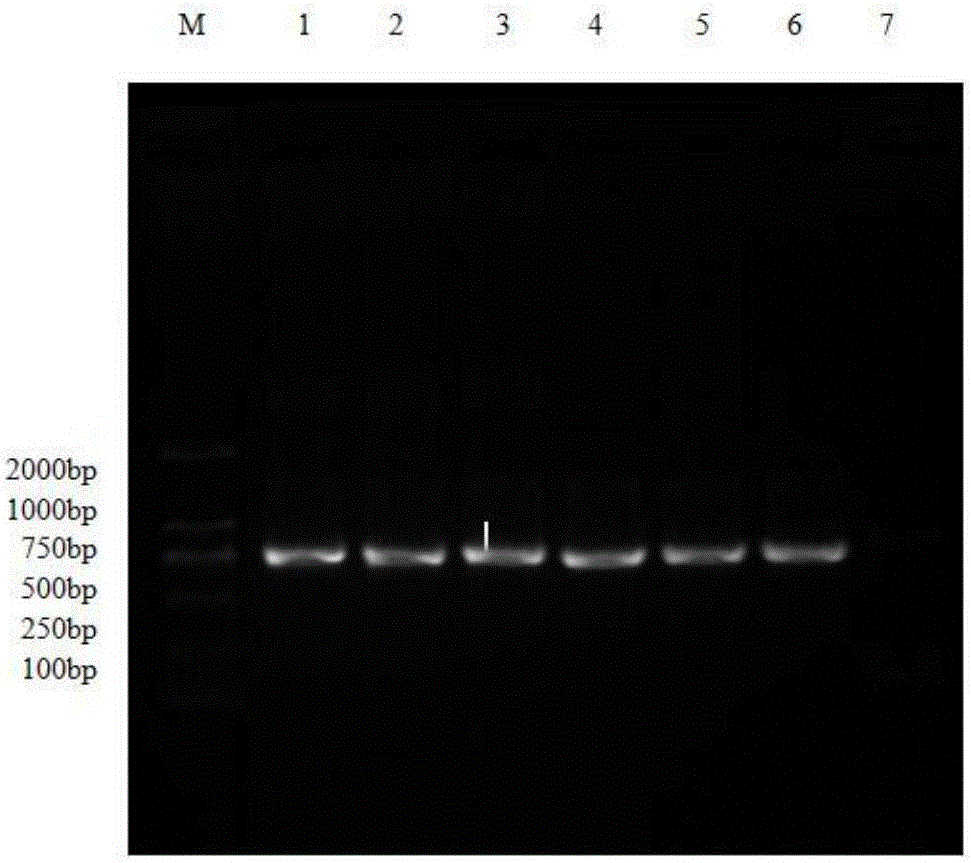
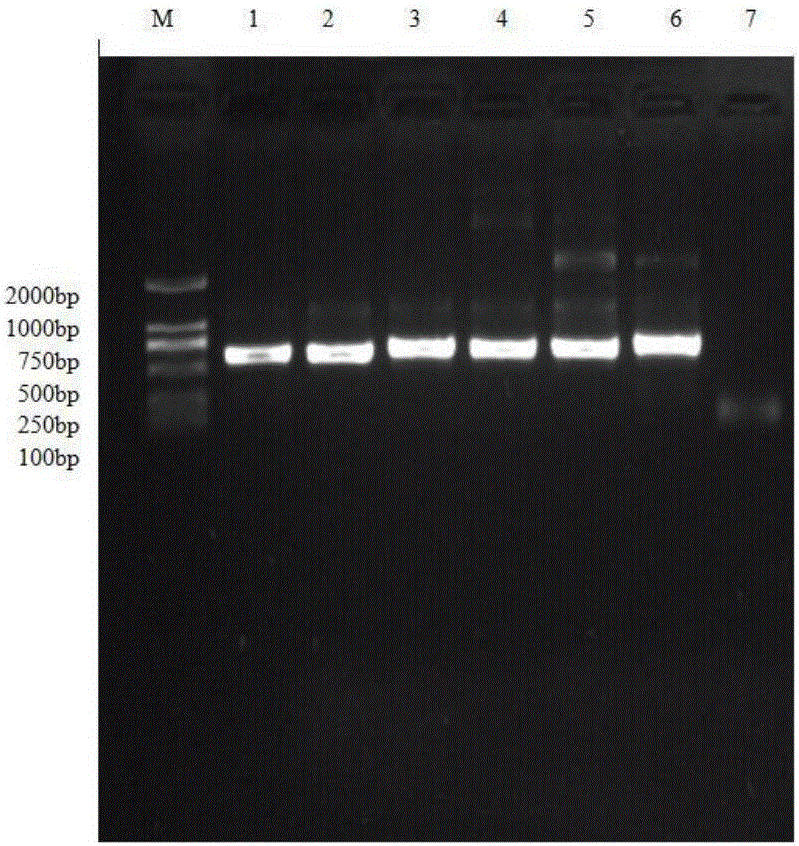
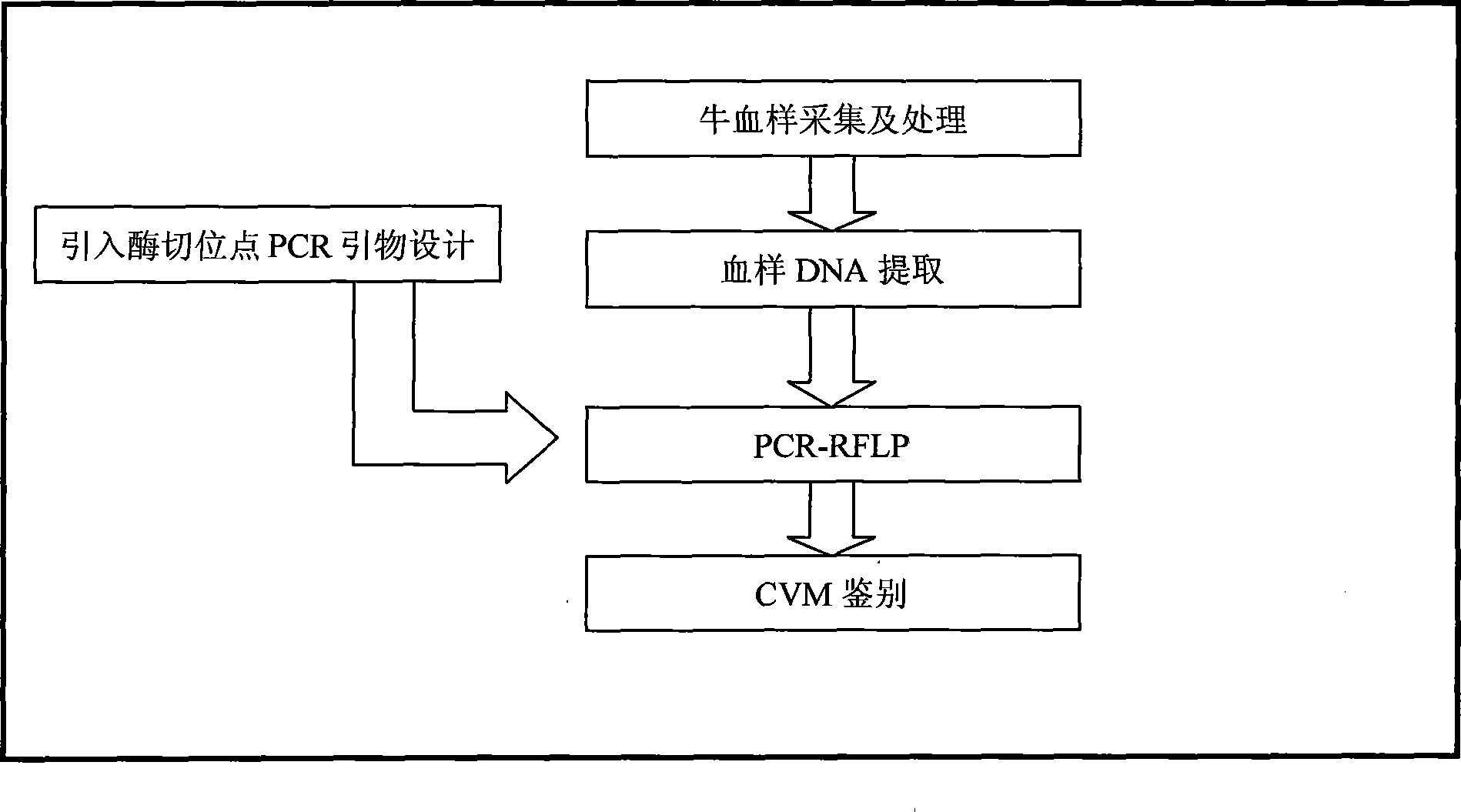
PCR-RFLP detection method for complex vertebral malformation of milk cow and crossbred cattle
The present invention belongs to the field of animal disease detection technology, specifically to the field of cattle recessive genetic disease spondyle malformation syndrome (CVM) detection, includes steps as follows: extracting cattle genome DNA from the cattle sample to be tested, designing the PCR amplification prime pair intruding the restriction enzyme site and performing the PCR reaction based on the cattle SLC35A3 gene sequence, using 2.5% agarose gel electrophoresis to test the PCR amplification products, dyeing, observing bands in the gel imaging system, recovering the PCR products to perform restriction enzyme, performing electrophoresis detection in 10% polyacrylamide gel after the reaction is completed, determining the genetype of the tested sample according to the band distribution of the amplification products.
Owner:HUAZHONG AGRI UNIV
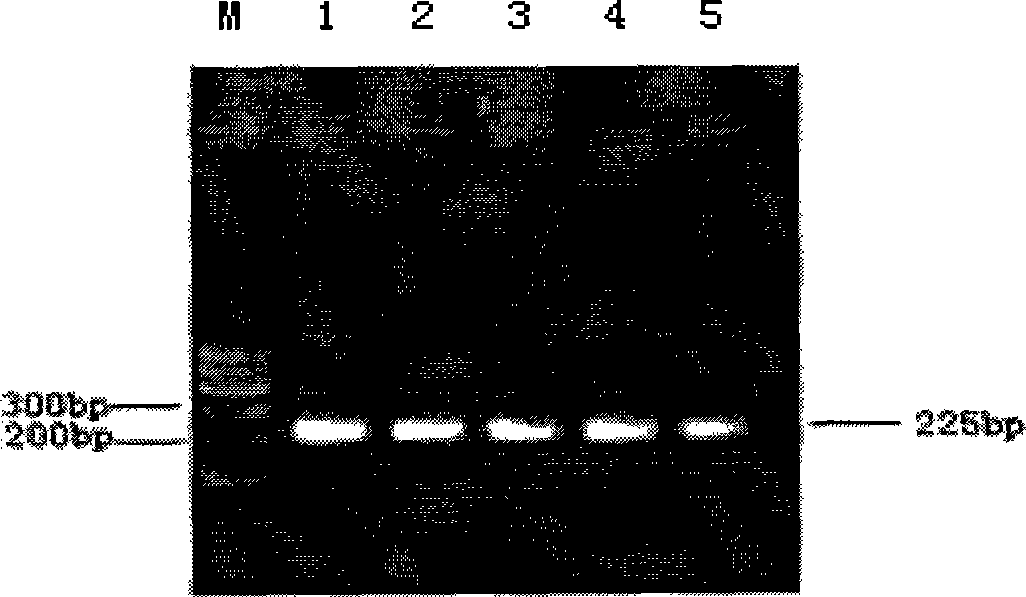
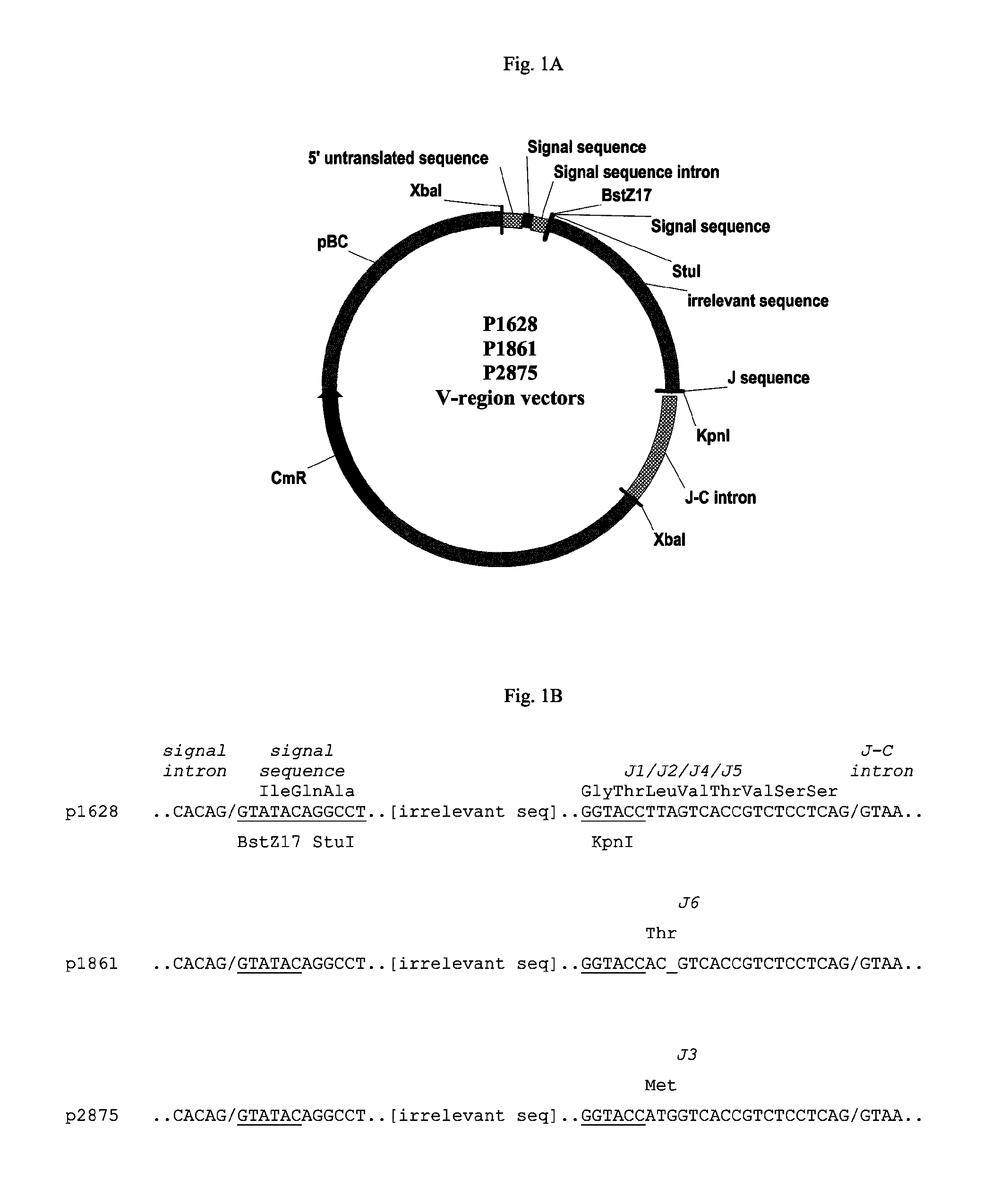
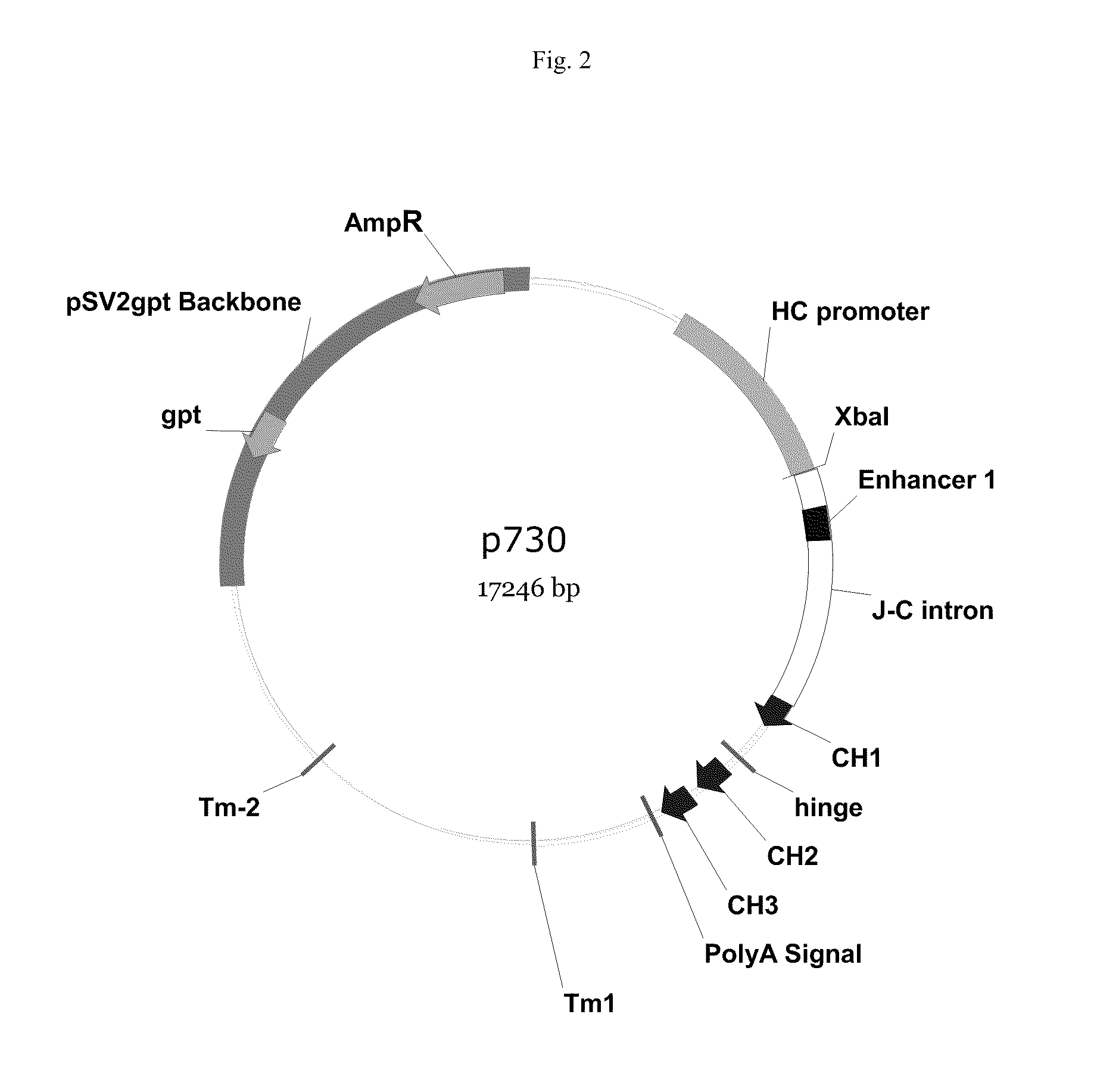
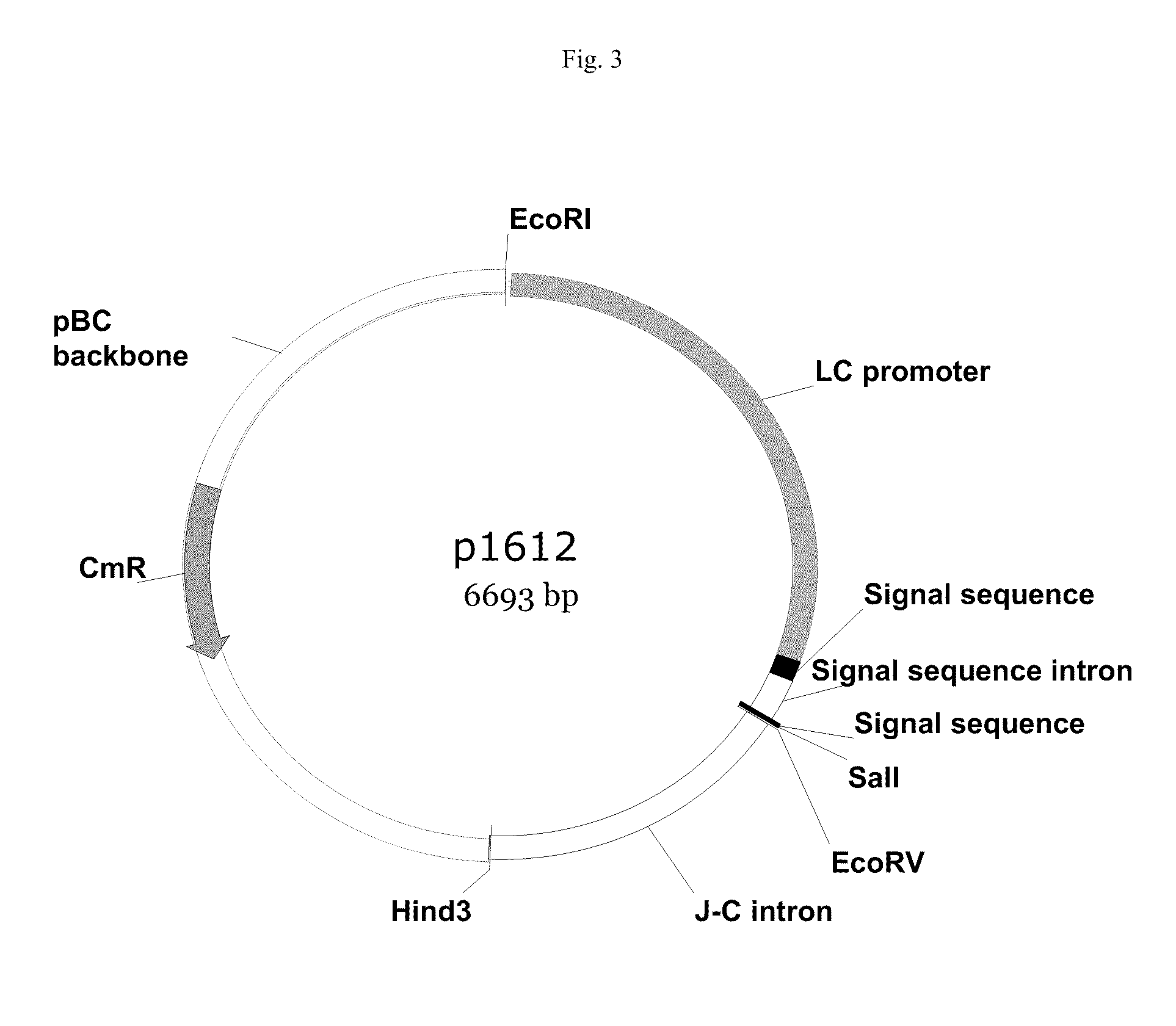
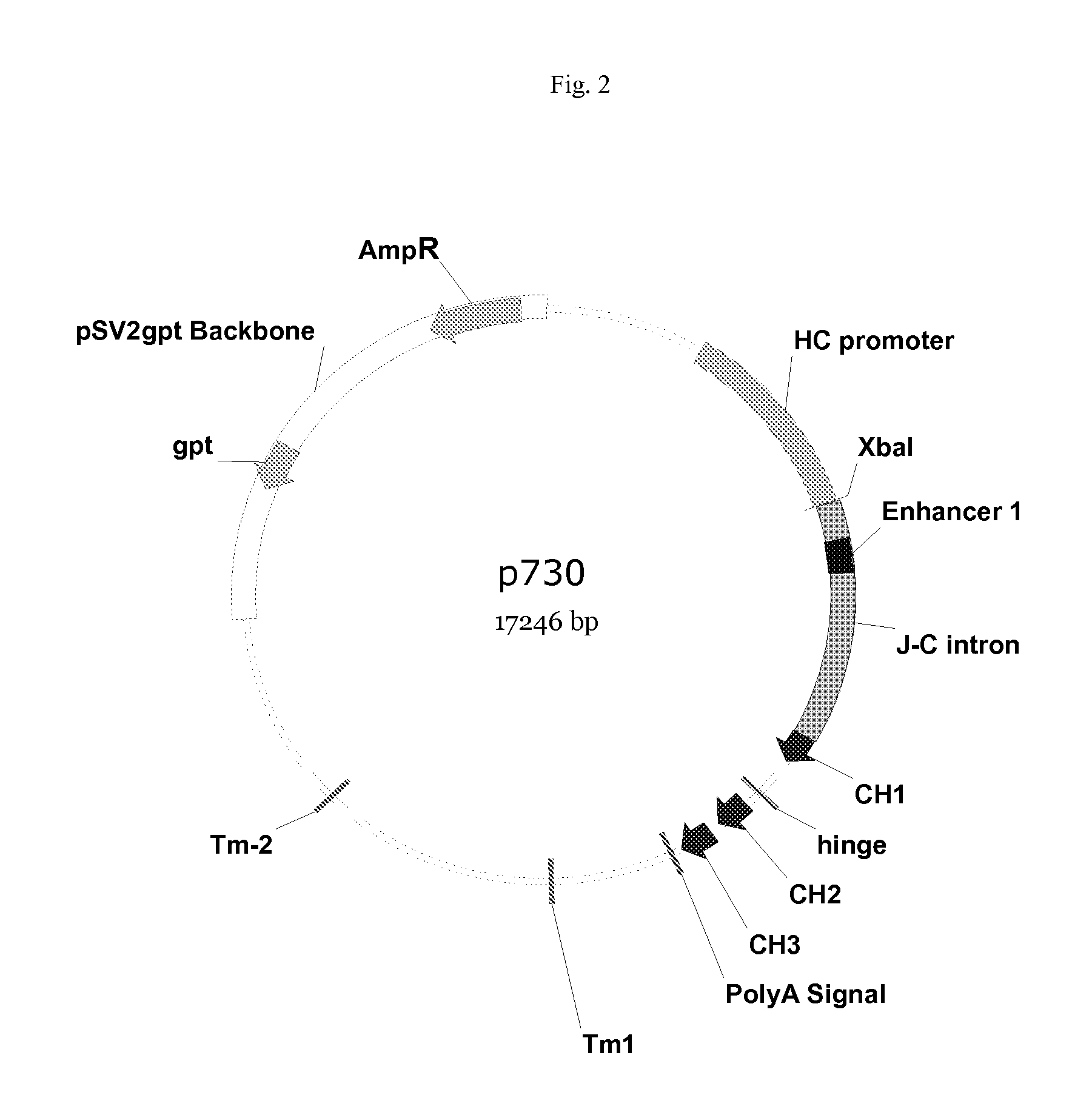
Vectors, host cells, and methods of production and uses
ActiveUS8663980B2Effective transcriptionIncrease yield and efficiencyAnimal cellsAntibody mimetics/scaffoldsAntibody expressionAntibody dna
Owner:JANSSEN BIOTECH INC
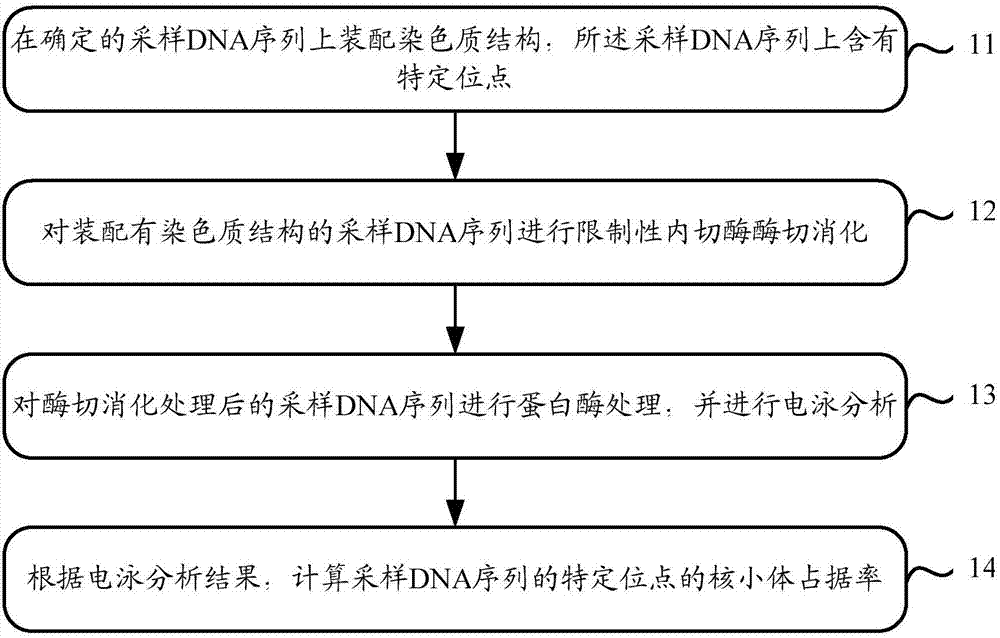
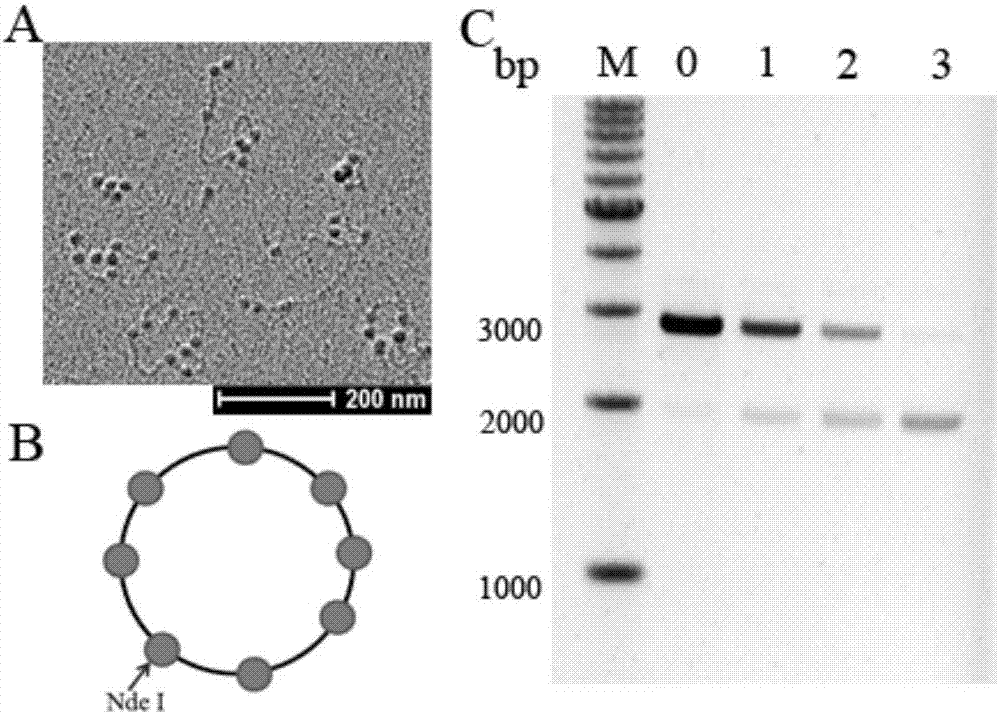
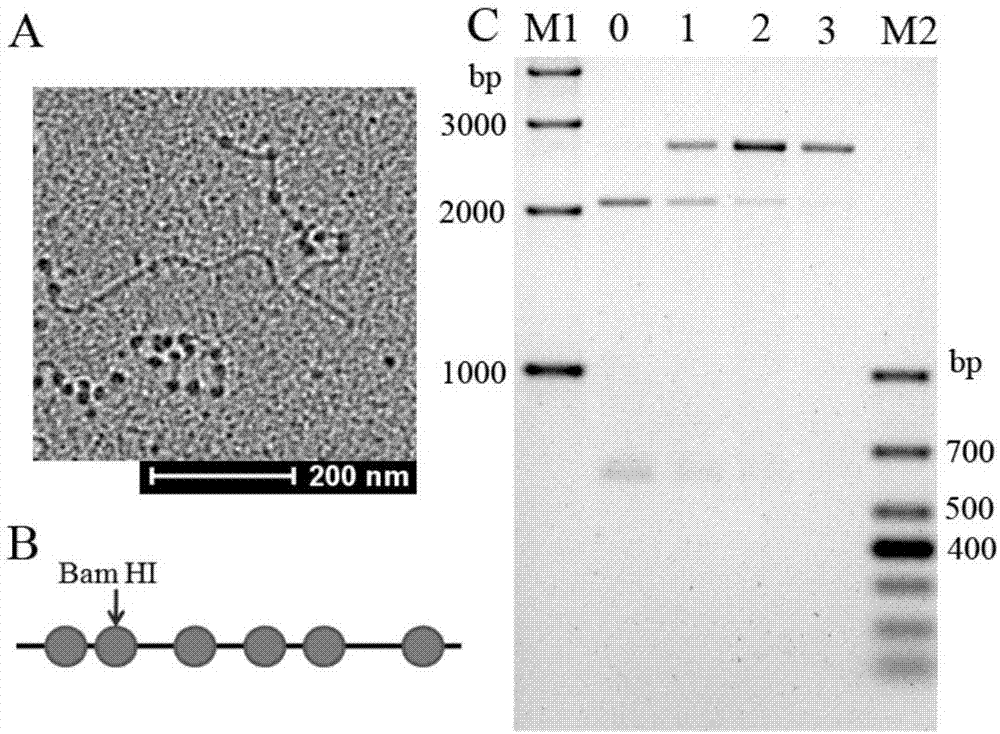
Capacity detection method for assembling chromatin structure on specific site of DNA sequence
ActiveCN107541558AThe implementation process is simpleThe realization process is convenientMicrobiological testing/measurementEnzyme digestionElectrophoresis
The invention relates to the field of molecular biology, in particular to a capacity detection method for assembling a chromatin structure on a specific site of a DNA sequence, and aims to solve the problem that an existing technology cannot detect the nucleosome occupying rate at a fixed site to determine the chromatin structure assembling capacity. The method is mainly to assemble the DNA sequence containing a specific restriction enzyme site into the chromatin structure; enzyme digestion is carried out through restriction enzyme to remove various types of protein, then electrophoresis detection is carried out, and the site and the nucleosome occupying rate of the DNA sequence near the site are quantitatively calculated to judge a local chromatin structure near the site. Therefore, the efficiency of assembling the chromatin structure on the specific site on the DNA sequence and the nucleosome assembling capacity are analyzed. The detection method is simple and quick in realizing process, and high in specificity and accuracy.
Owner:INNER MONGOLIA UNIV OF SCI & TECH
Preparation method of target sequence random sgRNA full-coverage group
The invention provides a preparation method of a target sequence random sgRNA full-coverage group. The preparation method comprises the following steps: cutting a PAM region of a target sequence by using restriction enzyme; connecting an upper sgRNA skeleton; obtaining a sequence of a targeting original spacer region through a side-cut activity restriction enzyme site on the sgRNA skeleton; connecting a T7 promoter; performing amplification to obtain an sgRNA library with a T7 promoter; and carrying out in-vitro transcription to obtain a target sequence sgRNA library. The invention also discloses a preparation method of the sgRNA library of the ribosomal RNA, a method for removing the ribosomal RNA in the RNA library, and a method for removing a human whole genome from a host genome. The method disclosed by the invention has the advantages of low cost, simplicity in manufacturing, uniform coverage, small preference, no limitation by the length of a target sequence, no need of designing sgRNA on a large scale and the like.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
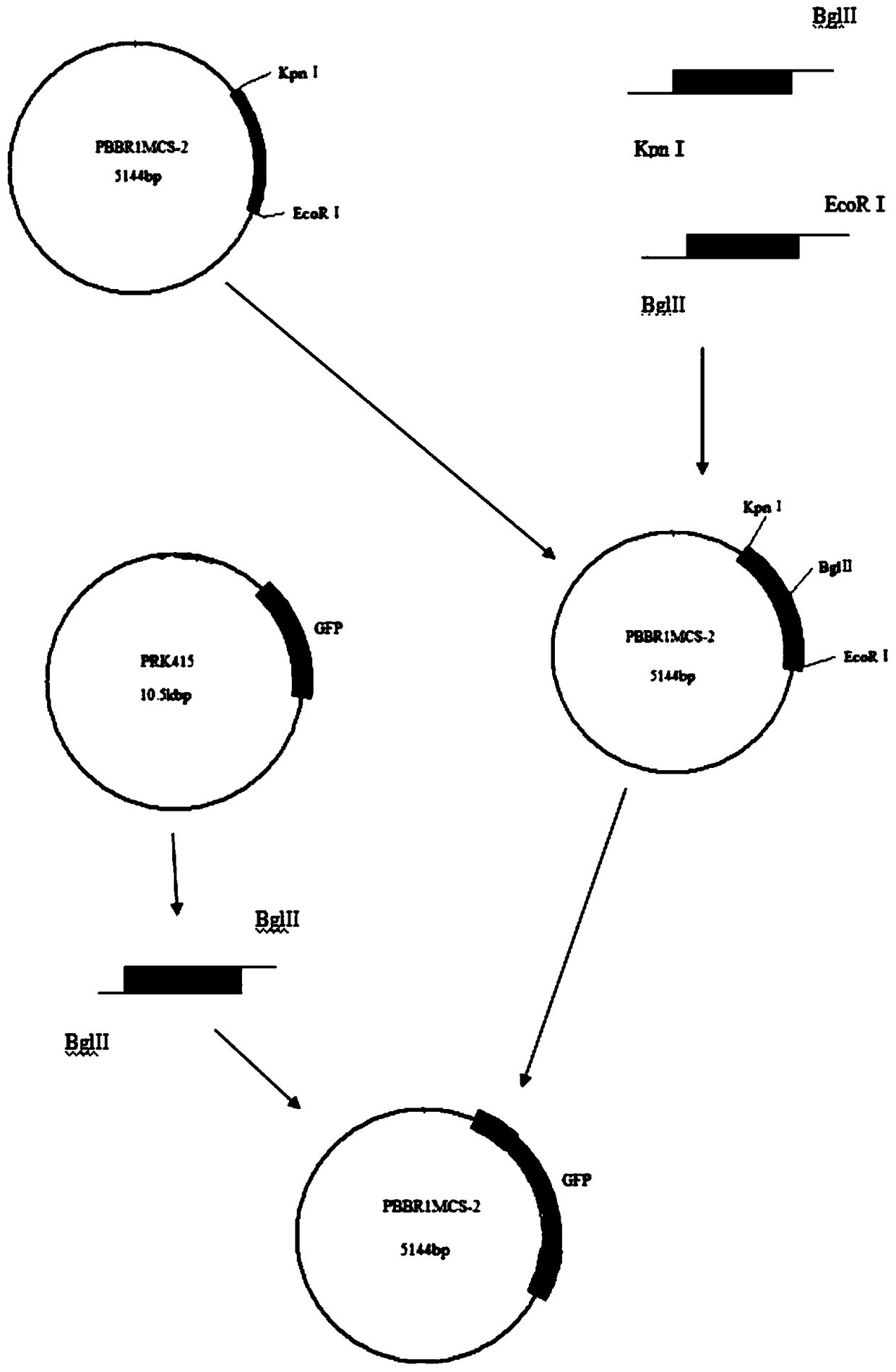
Construction and electrotransformation method of photosynthetic bacteria rhodopseudomonas palustris fluorescent GFP marker vector
InactiveCN108949792AStable fluorescence expressionObserving Colonization BehaviorBacteriaVector-based foreign material introductionShuttle vectorEnzyme digestion
The invention discloses a vector recombination and construction method, a photosynthetic bacteria rhodopseudomonas palustris fluorescent GFP marker vector construction and electrotransformation method. The recombinant vector includes the photosynthetic bacteria rhodopseudomonas palustris fluorescent GFP marker vector including a PBBR1MCS-2 shuttle vector and a full-length fragment of a green fluorescent protein marker gene GFP, and is fused with a photosynthetic bacteria genome expression promoter PpckA gene and terminator TpckA gene, as well as a widely applied GFP gene, and a full-length fragment sequence of the GFP gene; the full-length fragment of the GFP gene is located between two ECORI and SACI restricted restriction enzyme sites of the PBBR1MCS-2 shuttle vector. The construction method includes the steps: genome DNA extraction, primer designing, PCR amplification, enzyme digestion, connection and other steps. The invention also includes obtained engineering bacteria containingthe recombinant vector, wherein the engineering bacteria are obtained by electrotransformation of the recombinant vector into competent cells. The engineering bacteria have the advantages of high stability, simple technology and environmental friendliness, and can be applied for study of a colonization behavior of photosynthetic bacteria in plants.
Owner:HUNAN PLANT PROTECTION INST
Construction method of HIV-1 integrase mutant library
InactiveCN104975353AHigh resolutionImprove efficiencyVector-based foreign material introductionLibrary creationProtein targetRestriction site
The present invention discloses a construction method of an HIV-1 integrase mutant library. The method comprises the following elements: (1) manufacturing gaps on random sites of a target gene by using transposition of transposon and restriction enzyme on both ends of the transposon, inserting mutation insertion boxes, using the restriction enzyme sites on the box to achieve replacement of three bases on the gene through digestion self-connection, and translating the target gene into protein to cause mutation o one or two consecutive amino acids on the target protein; and (2), using site-directed mutagenesis PCR to realize mutation of three bases, for the replacement of the original target gene and on the inserted mutation insertion boxes, from codons encoding cysteine into codons encoding other 19 amino acids, thereby increasing the diversity of the final mutant library.
Owner:ZHEJIANG UNIV
Bran coat source peroxidase anti-tumor active fragment as well as preparation method and application thereof
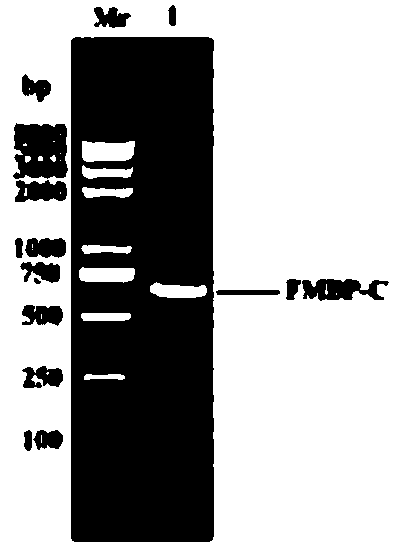
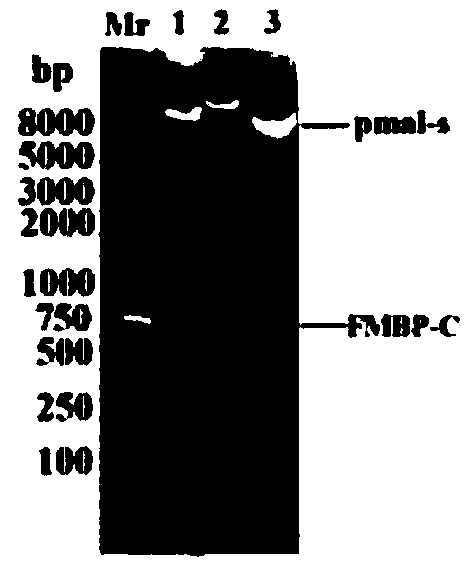
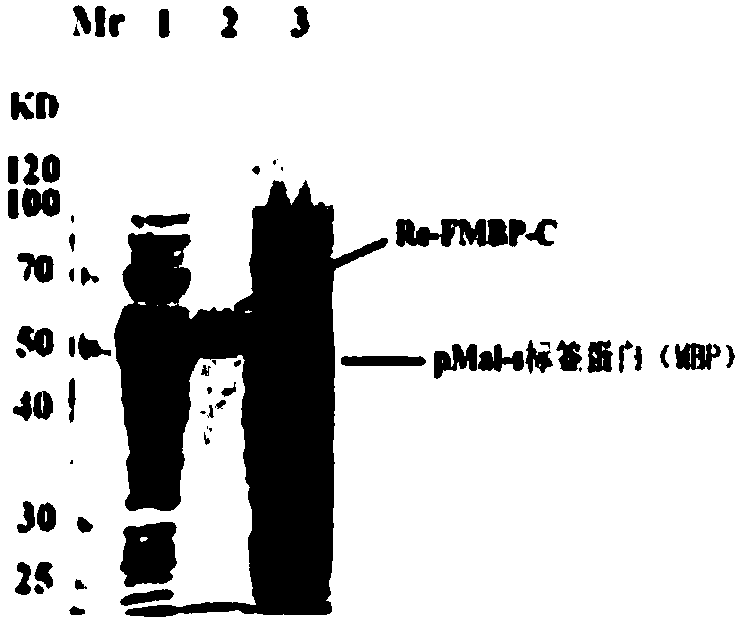
The invention provides a bran coat source peroxidase anti-tumor active fragment as well as a preparation method and application thereof. The preparation method of the fragment comprises the followingsteps: extracting mRNA of rice seedling tissues by a Trizol method, taking the mRNA as a template, performing enzymatic catalysis, and performing inverse transcription to obtain cRNA; obtaining a protein gene sequence in Genebank according to FMBP mass spectrum identification results of the rice anti-tumor active protein, respectively performing truncated screening according to different functional regions, determining a gene sequence of Ca<2+> binding sites as an anti-tumor effect structural domain, and designing a specific primer thereby; taking the rice cDNA as a template, and amplifying the FMBP-C gene sequence by a PCR (Polymerase Chain Reaction) method; respectively introducing BamH I and HindIII restriction enzyme sites at the upstream and downstream of the sequence, connecting thesites with a pMal-s vector, constructing a pMal-s-FMBP-C recombinant plasmid, transferring E.coli DH5alpha competent cells, and screening to obtain a positive transformant; performing induced expression on a target protein through IPTG (Isopropyl Thiogalactoside), performing affinity purification, desalting and concentrating, and detecting functions of resisting tumors and reversing multiple drugresistance of tumors by an MTT method. The results show that the active fragment has obvious activities of resisting tumors and reversing multiple drug resistance of the tumors.
Owner:SHANXI UNIV
Support for fixing nucleotide and process for producing the same
InactiveCN1396954AEffective generationEasy to fixSugar derivativesMicrobiological testing/measurementNucleotideRestriction site
A support for fixing a nucleotide which contributes to the efficient clarification of DNA without damaging the terminal parts of the DNA and, therefore, is useful in the fields of molecular biology, biochemistry and the like. This support for fixing a nucleotide is a chemically modified substrate on which oligonucleotides are immobilized characterized in that the oligonucleotides have a restriction enzyme cleavage site. This support is produced by chemically modifying the substrate, immobilizing a single-stranded oligonucleotide thereon, then hybridizing the single-stranded oligonucleotide with another single-stranded oligonucleotide having a base sequence complementary thereto, and ligating an oligonucleotide having a restriction enzyme site at the end thereof.
Owner:TOYO KOHAN CO LTD
Pre-T vector, preparation thereof and applications
The invention provides a pre-T-vector, which is prepared by the following method. (1) Plasmids of resistant genes of sensitive antibiotics of escherichia coli without containing ampicillin are taken as templates to conduct PCR amplification; XcmI restriction enzyme sites are introduced into terminals of a primer 5`; and DNA fragments are obtained by amplification which contain the resistant genes of the antibiotics, and the upstream and the downstream of the DNA fragment express regulation sequences, and the two terminals of the DNA fragment contain the XcmI restriction enzyme sites. (2) The DNA fragments obtained in procedure (1) are connected with a pUCmT vector by a joining enzyme T4DNA. (3) The connected product obtained in procedure (2) is transformed into escherichia coli DH5 Alpha or JM109; and positive clones are screened, and then the pre-T-vector can be obtained. The pre-T-vector has the following advantages that vectors prepared by the pre-T-vector can be used to clone the product PCR quickly, and X-gal is not needed in the process of screening the positive clones with low negative rate.
Owner:ZHEJIANG UNIV OF TECH
Recombinant lactobacillus rhamnosus engineering strain and preparation method thereof
InactiveCN101948856AImprove survival rateImprove antioxidant capacityBacteriaMicroorganism based processesEscherichia coliDismutase
The invention relates to a recombinant lactobacillus rhamnosus engineering strain and a preparation method thereof. The lactobacillus rhamnosus engineering strain carries expression vectors of a catalase gene and a superoxide dismutase gene which are connected in series. The method for preparing the engineering strain comprises the following steps of: 1) amplifying catalase gene segments and superoxide dismutase gene segments from genomes of lactobacillus sake and streptococcus thermophiles by a polymerase chain reaction (PCR) method; 2) designing a specific restriction enzyme site, wherein the restriction enzyme site is connected in series with an escherichia coli-lactobacillus shuttle plasmid vector pSIP502; and 3) transforming a recombinant plasmid into lactobacillus rhamnosus by an electroporation method. The recombinant lactobacillus rhamnosus engineering strain prepared by the method can remarkably enhance oxidation resistance and can be applied to the fermented food industry.
Owner:CHINA AGRI UNIV
Method for high-effective construction of T-carrier based on polymerase chain reaction
InactiveCN101177689ASimple methodShort cycleVector-based foreign material introductionPresent methodSingle strand
The invention discloses a method for efficiently constructing a T vector based on the PCR method. The main method is to introduce a pair of oligonucleotide fragments containing a specific restriction enzyme site into the starting vector (such as pUC18). The restriction enzyme site is characterized by The corresponding enzyme (such as EcoRV) can produce blunt ends, and the bases before and after the cutting point are T and A, respectively. Then use the enzyme to linearize the starting vector as a PCR template, design primers to generate a single strand through single primer PCR amplification or asymmetric PCR amplification, and then anneal the two single strands to form a T vector. The preparation of the T vector by the method is simple and fast, and the prepared T vector can efficiently clone the PCR product with an "A" tail, and its non-recombination background is almost zero.
Owner:TIANJIN MEDICAL UNIV
Vectors, Host Cells, and Methods of Production and Uses
ActiveUS20110008321A1High yieldImprove efficiencyAnimal cellsAntibody mimetics/scaffoldsAntibody expressionEukaryotic plasmids
Antibody expression vectors and plasmids can incorporate various antibody gene portions for transcription of the antibody DNA and expression of the antibody in an appropriate host cell. The expression vectors and plasmids have restriction enzyme sites that facilitate ligation of antibody-encoding DNA into the vectors. The vectors incorporate enhancer and promoter sequences that can be varied to interact with transcription factors in the host cell and thereby control transcription of the antibody-encoding DNA. A kit can incorporate these vectors and plasmids.
Owner:JANSSEN BIOTECH INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com