Chromatin conformation capturing method
A chromatin conformation and chromatin technology, applied in chemical libraries, material testing products, combinatorial chemistry, etc., can solve the problems of inability to directly use quantitative clinical samples, low effective information capture rate, lengthy experimental procedures, etc., so as to shorten the experimental time. , improve specificity and sensitivity, reduce the effect of background
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
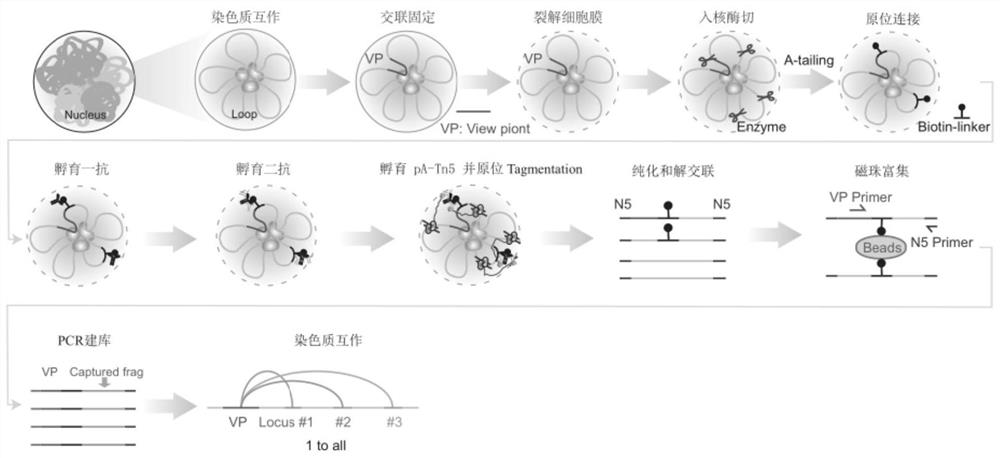
[0040] A chromatin conformation capture method (named as Tag-C), comprising the following steps:
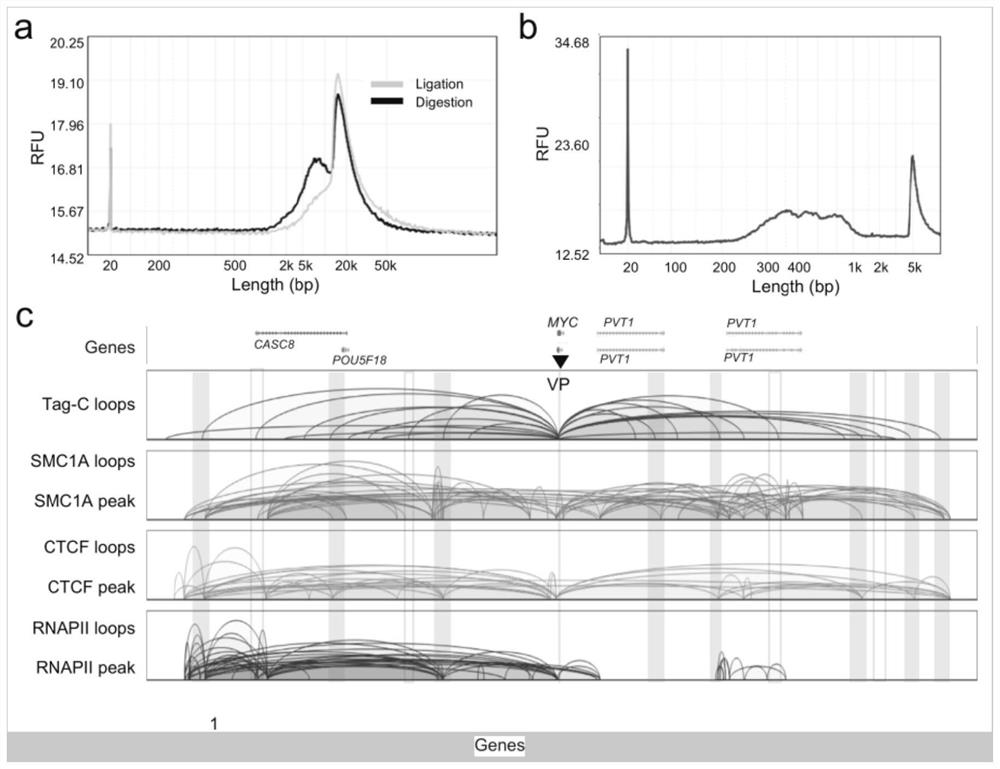
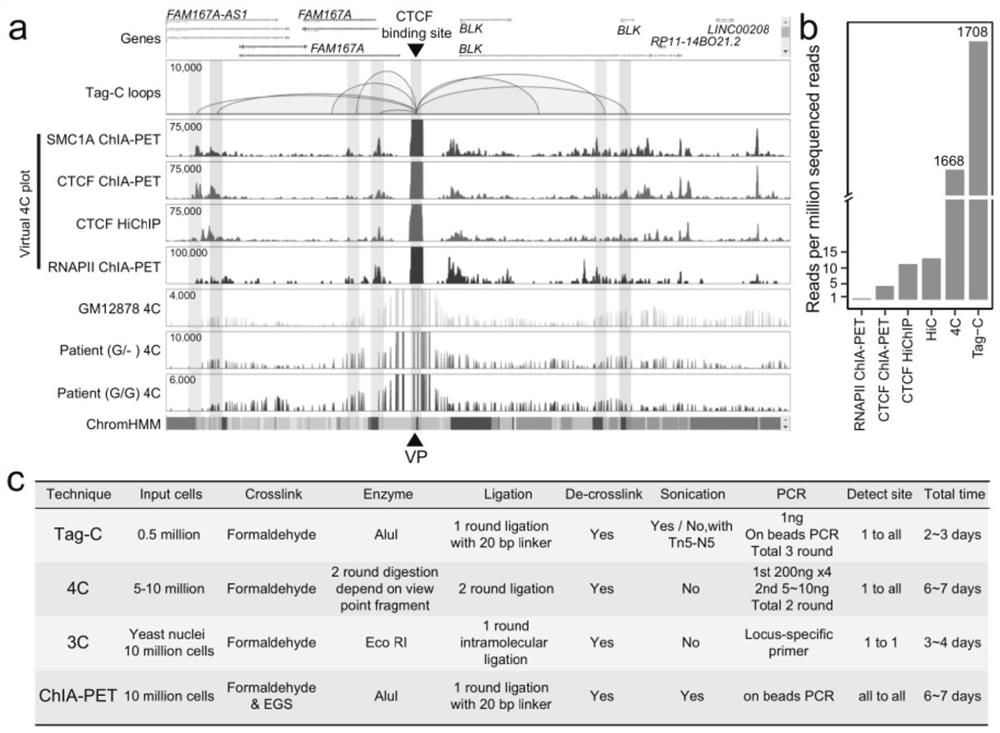
[0041] 1) Cell membranes were lysed with 0.1% SDS (sodium dodecyl sulfate) after cross-linking (cross-linking with 1% formaldehyde at room temperature for 10 minutes) to obtain cell nuclei, and nuclear membrane permeabilization was performed with 0.55% SDS ( Or use 0.05% TritonX-100 to perforate the cell membrane of uncrosslinked cells), use restriction enzymes (such as AluI, not limited to this) to fragment the chromatin DNA in situ in the nucleus to obtain blunt ends, Carry out the enzyme digestion reaction in a warm bath at 37°C for 1 to 2 hours, and the specific conditions are shown in Table 1;
[0042] 2) Add A bases (A-tailing) to the 3' end of the fragmented chromatin DNA with Klenow large fragments, 37°C for 1 hour, the specific conditions are shown in Table 2; Internally (in situ) use T4 ligase to combine the 3' end of the chromatin DNA close to the three-dimensional sp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com