Capacity detection method for assembling chromatin structure on specific site of DNA sequence
A DNA sequence and specific site technology, applied in the field of molecular biology, can solve the problems of cumbersome experimental steps, high experimental operation requirements, and high price, and achieve the effect of high specificity, high accuracy and simple implementation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
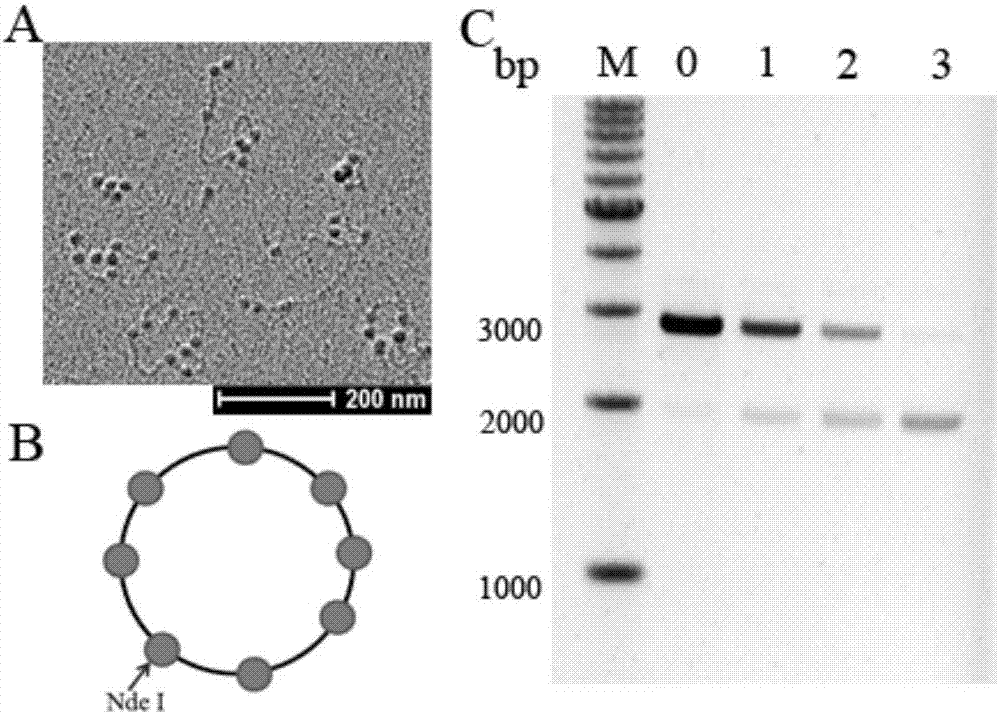
[0060] Example 1: Detection of ability to assemble nucleosomes at specific sites of reconstituted chromatin on circular plasmids (1) Reconstituted chromatin structure on circular plasmids. The purified circular plasmid pUC601 was mixed with the purified histone octamer as shown in Table 1, respectively, and dialyzed in TE solution containing 2 mol / L NaCl. During the dialysis process, TE was pumped into the dialysate to dilute the NaCl concentration in the dialysate to 0.6 mol / L, and the dialysis process was controlled for 16 hours; the dialysate was replaced with TE buffer without NaCl and continued dialysis for 3 hours. Wherein, the reaction temperature of the whole experiment process was controlled at 4°C.
[0061]
[0062] Table 1
[0063] (2) Restriction endonuclease digestion. The nucleosome occupancy near the Nde I single restriction site on the pUC601 plasmid sequence was detected. Take 1 / 5 of the dialyzed sample, add 8U of Nde I, and digest at 37°C for 1 hour.
...
example 2
[0066] Example 2: Detection of the ability to assemble nucleosomes at specific sites in remodeled chromatin on linear DNA sequences
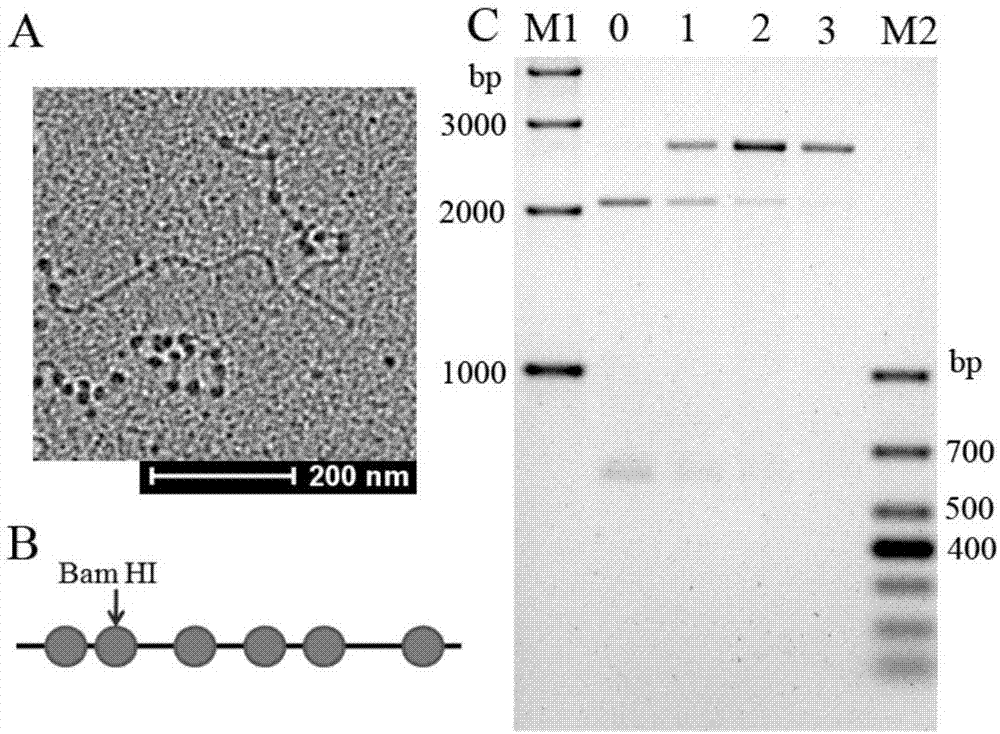
[0067] (1) Reconstruction of chromatin structure on linear DNA sequence. Plasmid pUC19 was digested with restriction endonuclease Ssp I into a linear DNA sequence. Mix the digested and purified linear plasmid pUC601 with the purified histone octamer as shown in the table below, and dialyze in TE solution containing 2mol / L NaCl. During the dialysis process, TE was pumped into the dialysate to dilute the NaCl concentration in the dialysate to 0.6 mol / L, and the dialysis process was controlled for 16 hours; the dialysate was replaced with TE buffer without NaCl and continued dialysis for 3 hours. The whole experimental process was controlled at 4°C.
[0068]
[0069] Table 2
[0070] (2) Restriction endonuclease digestion. The nucleosome occupancy near the EcoR I single restriction site on the pUC601 sequence was detected. Take 1 / 5 of the d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com