Quinoa Dimorphic Indel Molecular Marker and Its Development Method and Application
A molecular marker, quinoa technology, applied in biochemical equipment and methods, recombinant DNA technology, DNA / RNA fragments, etc., can solve problems such as confusion, lower genotyping accuracy, inconvenient use, etc., to reduce reading bands Error rate, ease of use, and abundant effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] The experimental methods in the following examples are conventional methods unless otherwise specified, and the involved experimental reagents and materials are conventional biochemical reagents and materials unless otherwise specified.
[0023] 1. Acquisition of quinoa genome sequence
[0024] The aerial parts of 11 quinoa germplasms at seedling stage were collected for DNA extraction. Genome sequencing libraries were constructed according to the operating instructions provided by Illumina. Genome Paired-end sequencing was performed using the Illumina HiSeq2500 sequencing platform of Berry and Kang Company, as shown in Table 1.
[0025] Table 1 The quinoa genome sequencing germplasm and its sequencing information
[0026]
[0027]
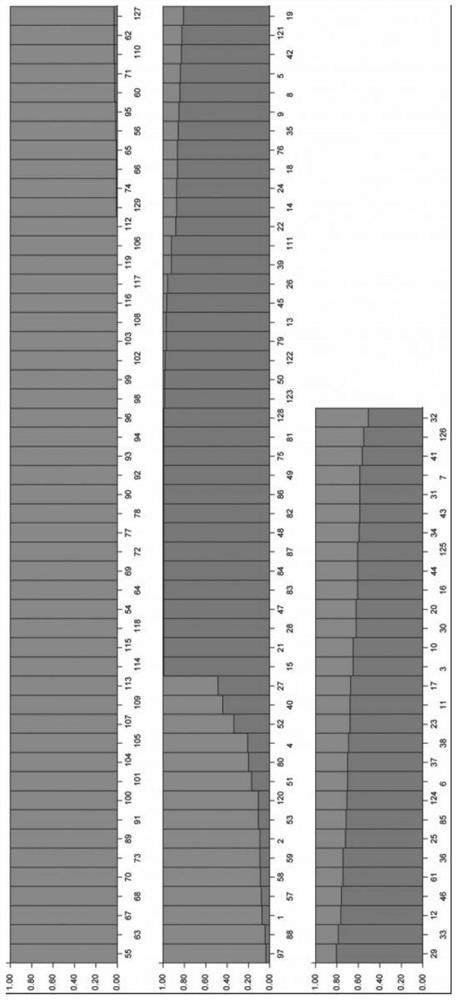
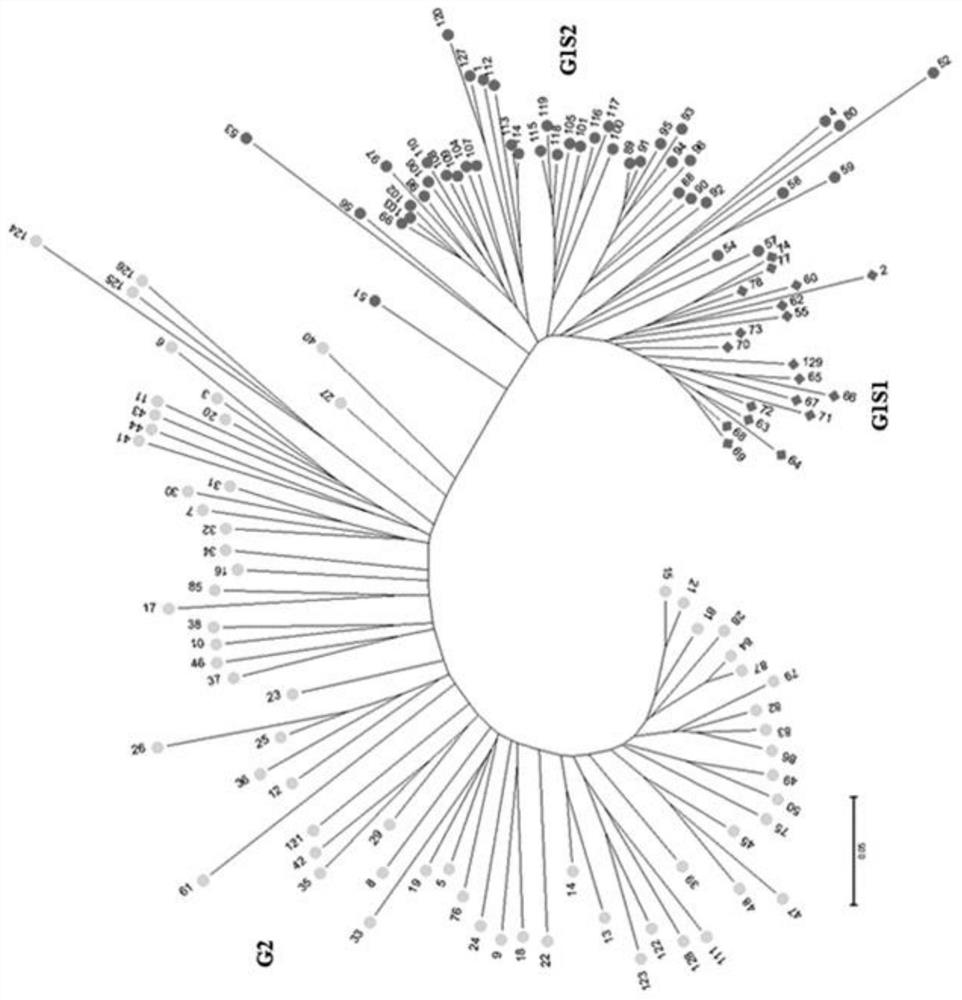
[0028] 2. Prediction and verification of quinoa dimorphic InDel molecular markers
[0029] The quinoa genome sequencing data (FASTQ format) was imported into mInDel software, and after quality control of the sequencing data, the hi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com