Visualizing modified nucleotides and nucleic acid interactions in single cells
A single nucleotide polymorphism and oligonucleotide technology, applied in the field of oligonucleotide probes for single nucleotide polymorphism detection and single nucleotide polymorphism detection, can solve the problem of reduced binding force, Can not hybridize and other problems, to achieve the effect of simple and cheap synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
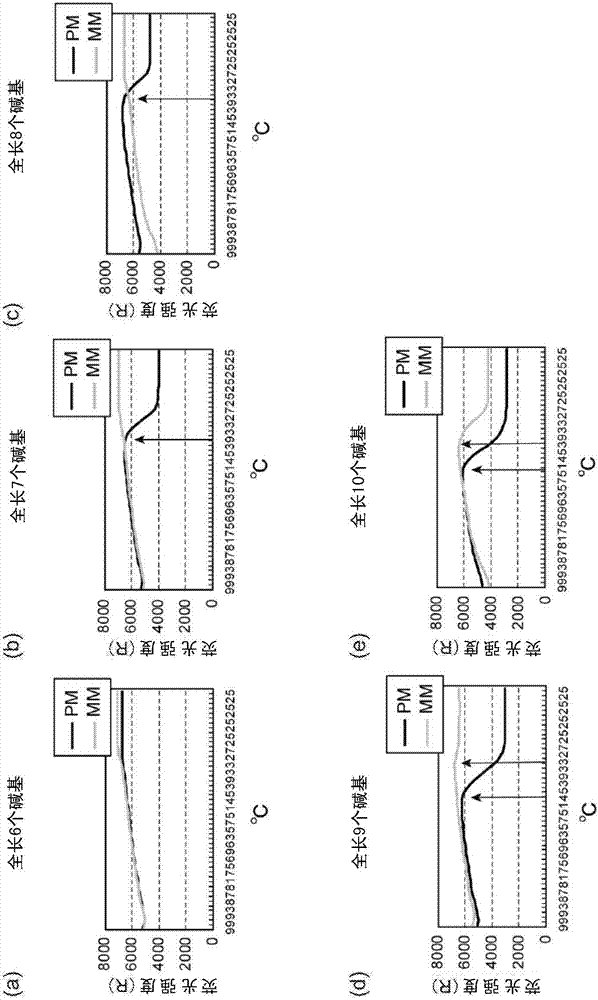
[0069] [Example 1: Effect of adding joint area]
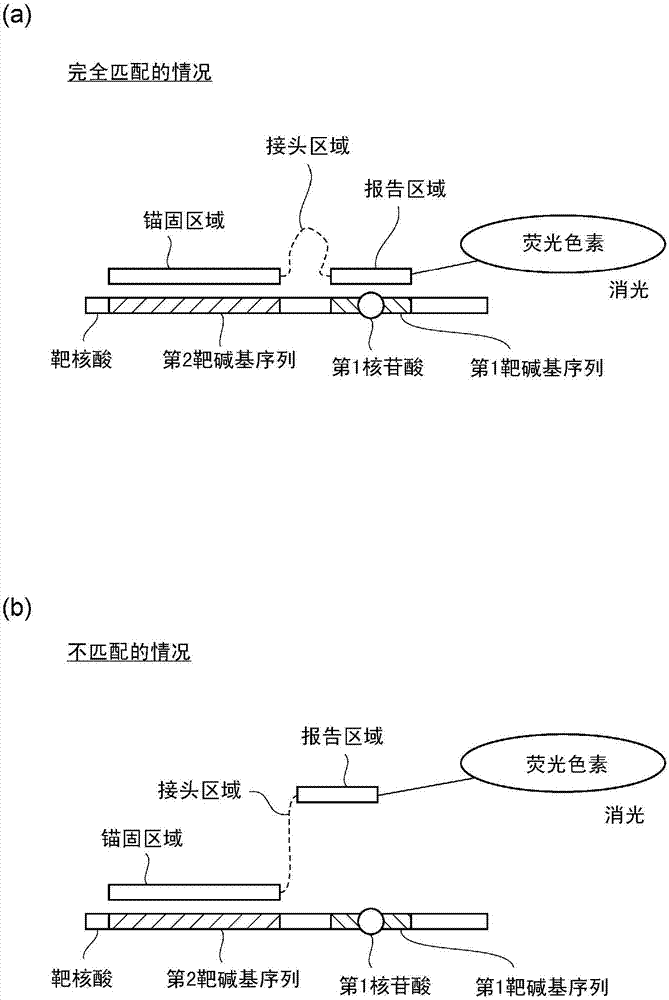
[0070] The effect obtained by interposing the reporting area and the anchoring area through the joint area was verified. As the detection system, Mycobacterium Tuberculosis is used to detect the missense variation of No. 516 Asp (GAC) in the rpoB gene replaced by Val (GTC). The change in fluorescence value caused by the hybridization of SNP-oligo DNA or linker-type SNP-oligo DNA with stepwise change in length to QProbe-bound DNA was analyzed. Since QProbe binds DNA with SNPs, use a system that can detect SNPs when there is a perfect match for SNP-oligo DNA or adapter-type SNP-oligo DNA, but cannot detect SNPs when there is no match.
[0071] (Material)
[0072] QProbe-bound DNA: DNA of QProbe-3G (manufactured by Nippon Steel & Sumitomo Metal Environmental Co., Ltd.) bound to DNA having a base sequence of SEQ ID NO: 1.
[0073] SNP-oligo DNA: DNA having any one of the base sequences of SEQ ID NOS: 2 to 11.
[0074] Adapter-t...
Embodiment 2
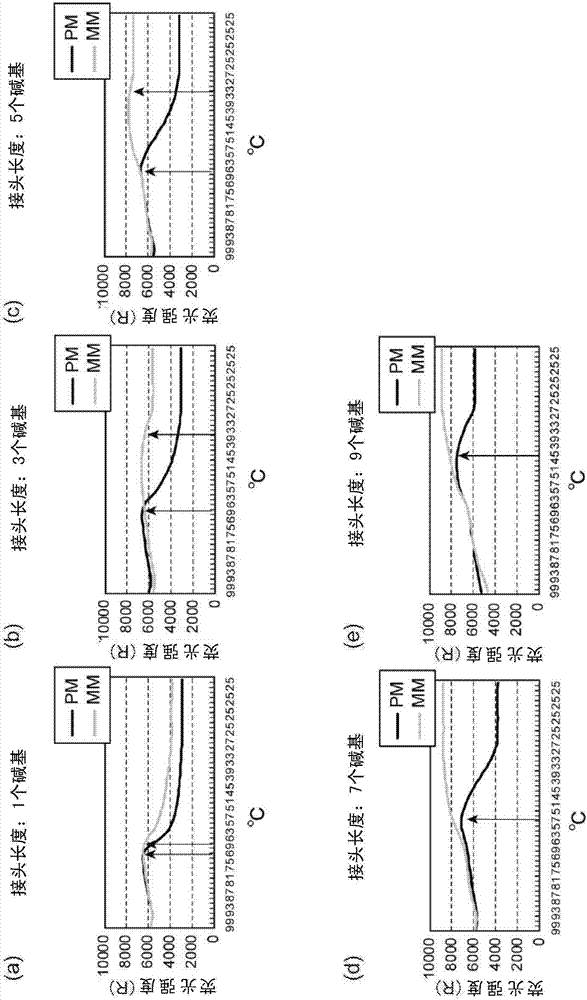
[0091] [Example 2: Study of Linker Region]
[0092] When designing QProbe binding probes, investigate the sequence of the anchor region and the length of the linker region used to hybridize the probe to the target nucleic acid.
[0093] (Material)
[0094] QProbe-bound DNA: DNA having a base sequence of SEQ ID NO: 22 to which QProbe is bound.
[0095] Linker type SNP-oligo DNA: It is an oligo DNA including a reporter region consisting of 4 bases and an anchor region consisting of 20 bases, based on DNA having a base sequence of SEQ ID NO: 23 or 24, Available in various lengths of spacers and joints. The reporting region is the 37th to 40th nucleotide sequence in SEQ ID NO: 23 or 24, and the anchoring region is 20 bases located on the 5' side from the reporting region by an interval of bases. The interval mentioned here refers to the number of bases from the 5' end of the reporter region to the 3' end of the anchor region when the reporter region and the anchor region are se...
Embodiment 3
[0101] [Example 3: Hybridization with synthetic DNA using an adapter-type QProbe binding probe]
[0102] Using the linker-type QProbe binding probe designed based on the results in Example 2, hybridization was performed with various synthetic DNAs to study the ability to identify SNPs.
[0103] (Material)
[0104] Synthetic DNA: DNA that has the base sequence of sequence number 23, 25, 27 or 29 and is completely matched with the adapter type QProbe binding probe, or has the base sequence of sequence number 24, 26, 28 or 30 and is compatible with the adapter type QProbe Binding to DNA that does not match the probe.
[0105] Adapter-type QProbe-binding probe: A probe in which QProbe is bound to nucleotides having any one of the nucleotide sequences of SEQ ID NO:31-34.
[0106] Hybridization buffer: 50 mM KCl, 10 mM Tris-HCl (pH 8.0), 0.1% Tween-20.
[0107] Table 4 shows the characteristics of the linker-type QProbe binding probe. In the sequences in the table, the underline...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com