Prediction method of drug release from pH-sensitive block copolymer micelles
A technology of block copolymers and prediction methods, applied in the directions of drugs or prescriptions, molecular design, etc., to achieve the effect of easy mechanism explanation and good fitting ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] This example aims to propose a method with a good degree of fit that can be used to accurately predict the release of amphiphilic pH-responsive block copolymer micelles near tumor tissue at different time periods. According to the complex structure of the block copolymer, the net drug release in different periods after self-assembly into drug-loaded micelles can be predicted, and then the drug release performance of the target block copolymer in acidic environment can be predicted And evaluation, to provide theoretical support for the realization of the goal of stable product quality and rapid response to market demand in industrial production.
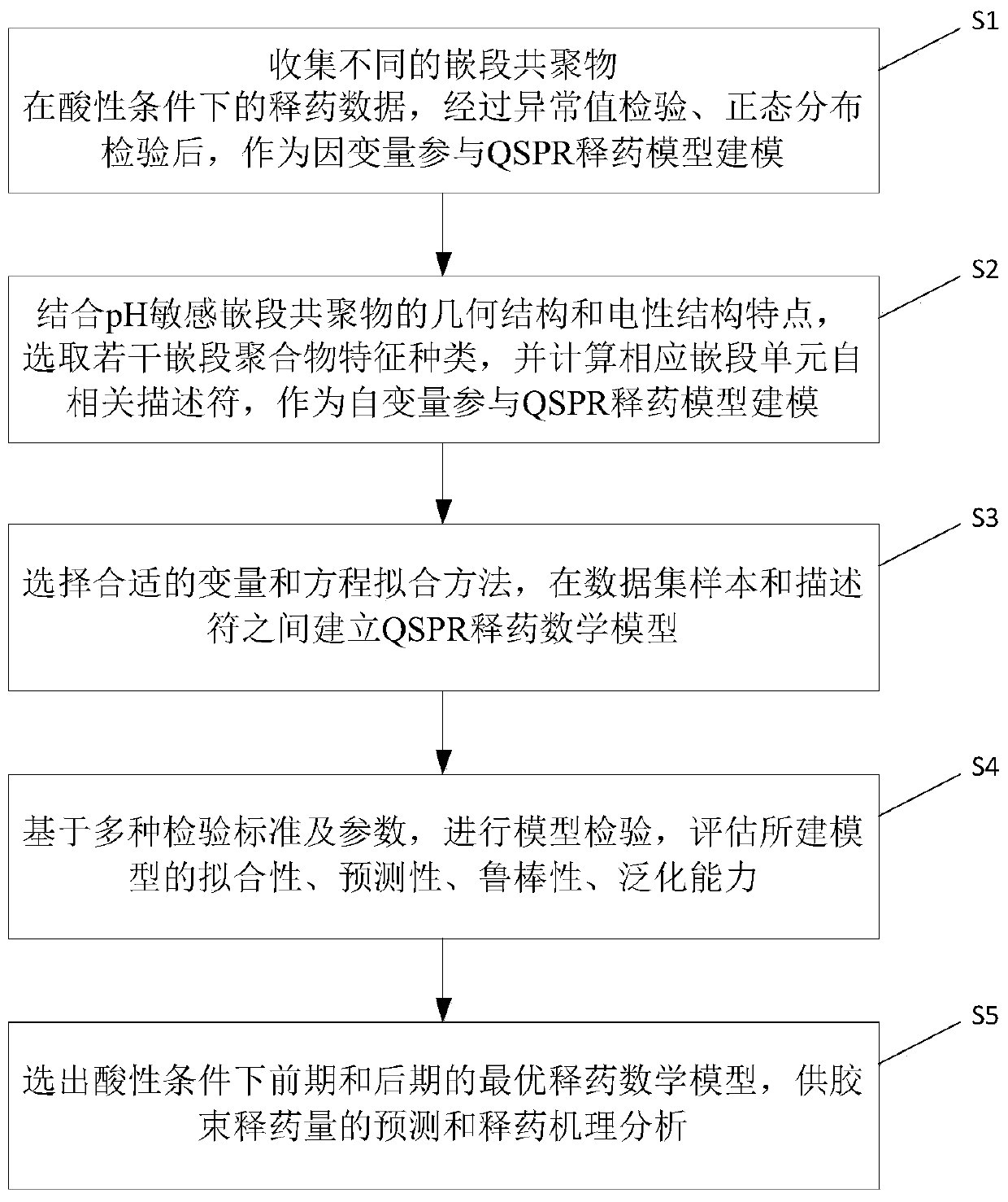
[0063] as attached figure 1 As shown, the method for predicting the drug release of the pH-sensitive block copolymer micelles disclosed in this embodiment comprises the following steps:
[0064] S1. Prepare reliable data set samples. The drug release data of different block copolymers under acidic conditions (pH=5.0) were col...
Embodiment 2
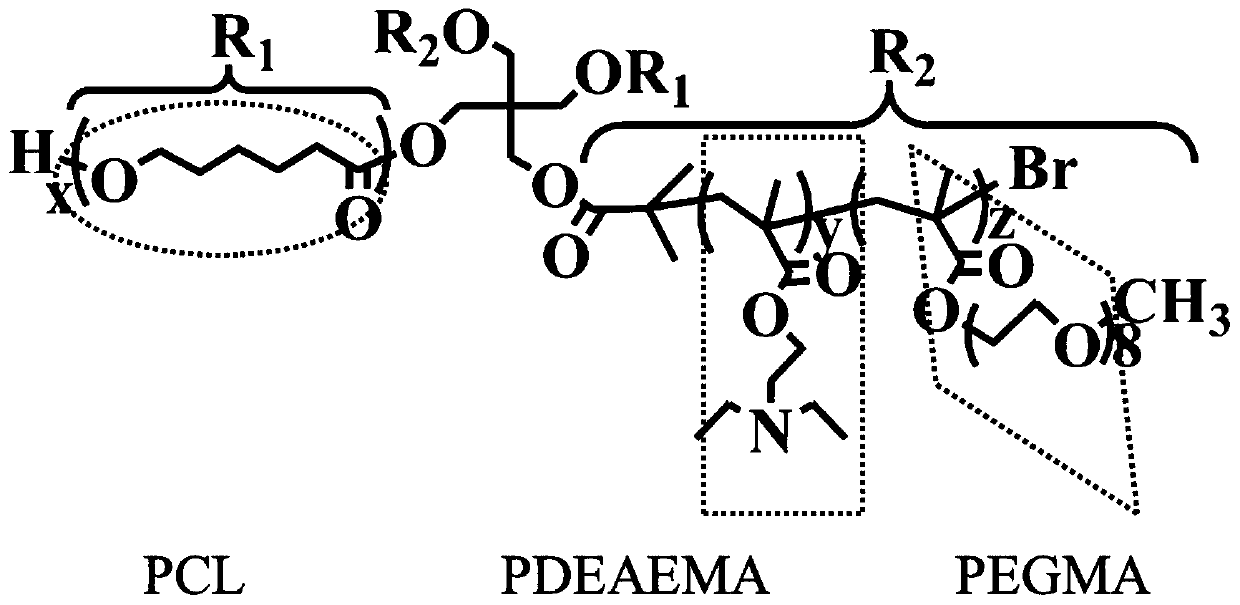
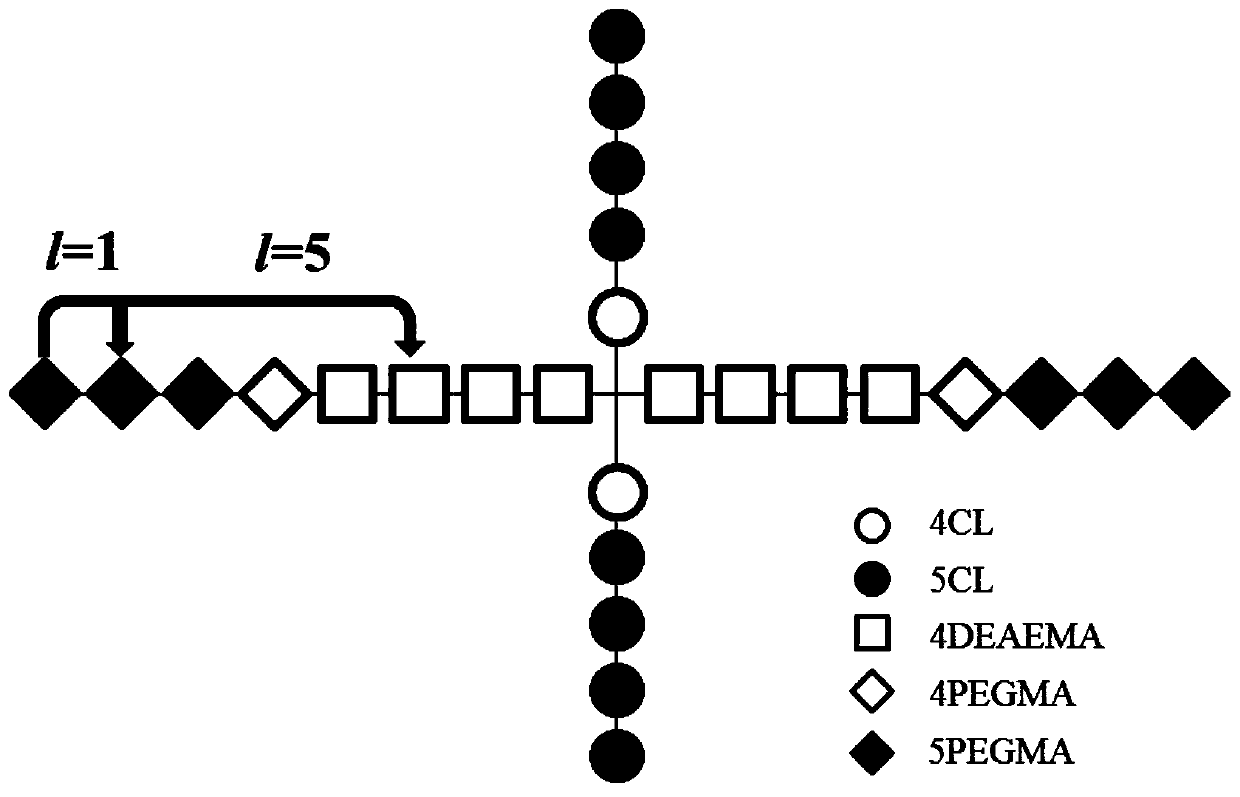
[0113] Given a tetraheteroarm block copolymer (PCL 24 ) 2 (PDEA 37 -b-PPEGMA 15 ) 2 , to predict its drug release in the early and late stages. First of all, based on the three-dimensional structure of the block copolymer, use Material Studio 7.0 software to optimize its structure and minimize its energy, and the values of 10 characteristics can be calculated. Then it is divided into several block units according to its topology, and the corresponding BUA descriptors with different step lengths are calculated, and the values of LogP(1), LogP(4), LUMO(4)gap(8) are 1.374, 1.4834, 1.604 respectively. , -1.764, and the values of LUMO(2), LUMO(6), and dipole(8) are 0.901, 1.075, and 1.317, respectively. Then, through the obtained application domain characterization diagram, it can be concluded that the compound falls within the scope of the application domain, so this model can be used for prediction.
[0114] Substitute the values of the above descriptors into the up...
Embodiment 3
[0121] Given a hexaheteroarm block copolymer (PCL 33 ) 3 (PDEA 12 -b-PPEGMA 10 ) 3 , it is necessary to predict its drug release in the early and late stages. The geometric structure of this polymer was constructed in Material Studio software, its structure was optimized and its energy was minimized, and 10 kinds of eigenvalues were calculated. Then divide its two-dimensional topological structure into different types of block units, and calculate the BUA descriptors with different step lengths. The values of LogP(1), LogP(4), LUMO(4)gap(8) are respectively - 1.446, -1.708, 0.957, 0.867, while the values of LUMO(2), LUMO(6), and dipole(8) are 0.166, 0.337, 0.665, respectively. Depend on Figure 5 From the Williams diagram drawn in , it can be seen that the compound falls within the scope of the application domain, so the pre-late model can be used to predict the drug release of this block copolymer.
[0122] Substituting the values of the above descriptors into ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com