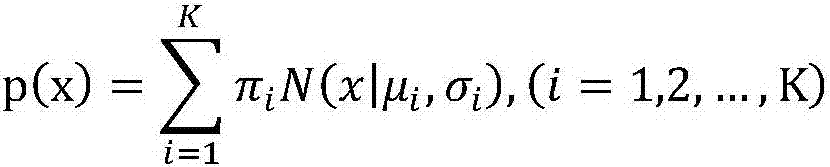Method for acquiring concentration of fetal cell-free DNA
A fetal and concentration technology, applied in the direction of instruments, calculations, electrical digital data processing, etc., can solve the problems of the influence of the accuracy of the results, the inability to dynamically obtain the statistical interval, etc., and achieve the effect of ensuring accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
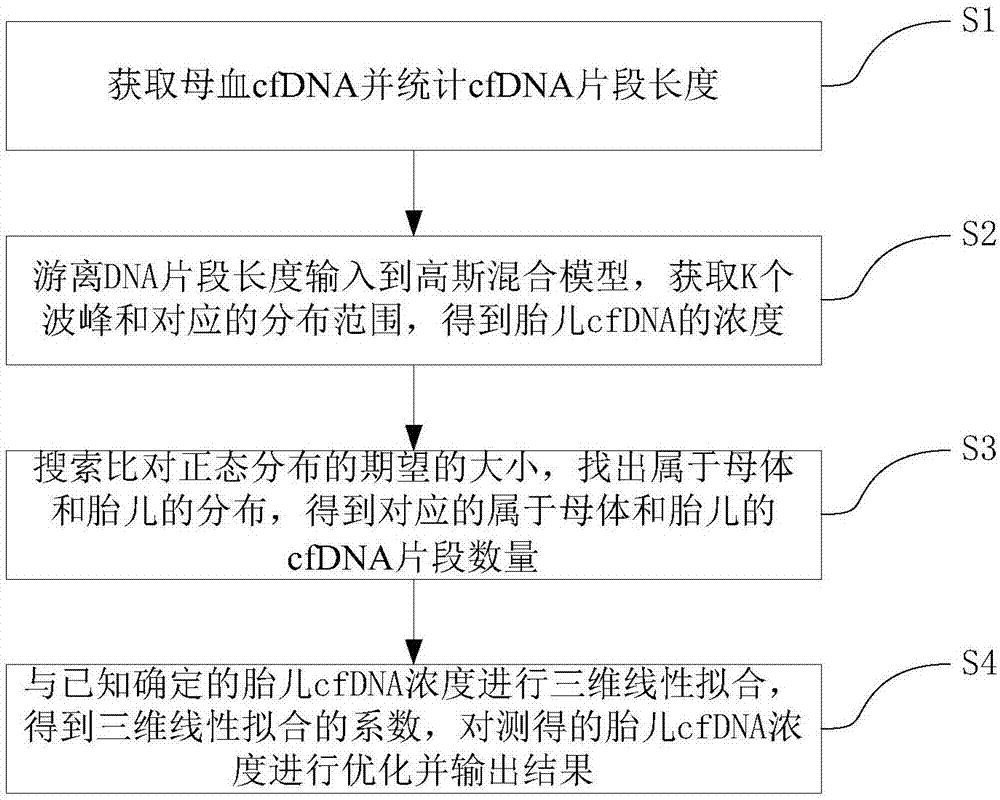
[0033] Such as figure 1 As shown, a method for obtaining fetal cell-free DNA concentration comprises the following steps:
[0034] S1: Obtain cell-free DNA (cfDNA) from maternal blood, and count the length of cfDNA fragments;
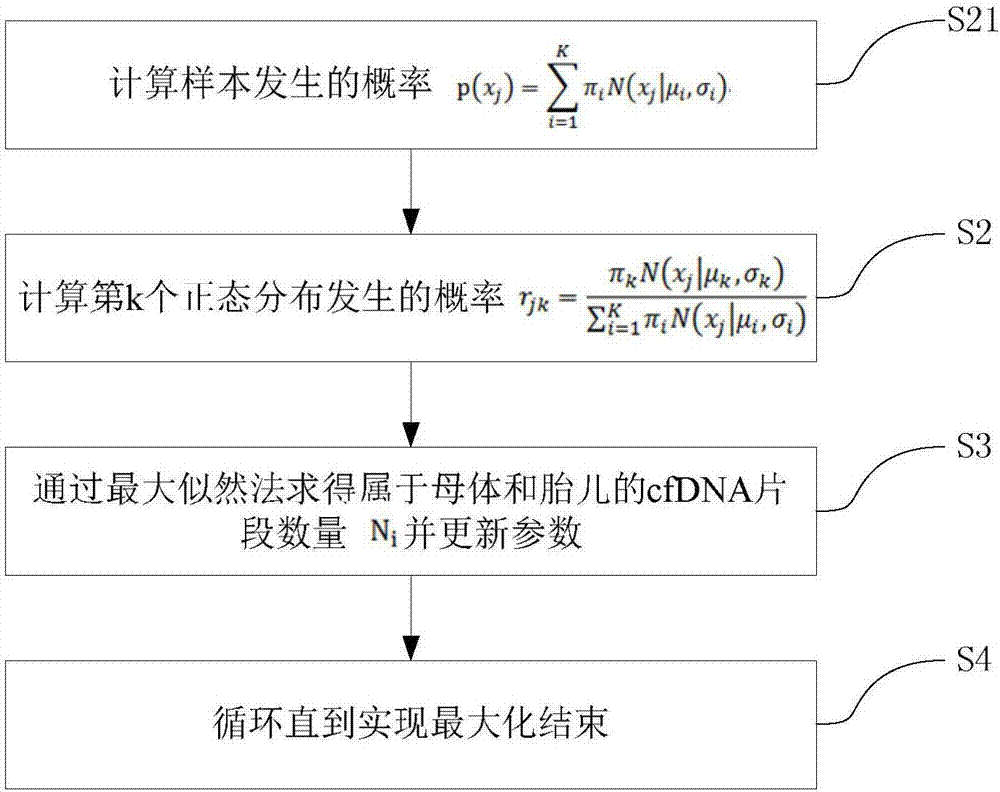
[0035] S2: Input the statistical cell-free DNA fragment length into the Gaussian mixture model, and use the Gaussian mixture model with K normal distributions to quantify the cell-free DNA of the mother and fetus, obtain K peaks and corresponding distribution ranges, and obtain fetal cell-free DNA concentration;
[0036] S3: By searching and comparing the expected size of the normal distribution, find the distribution belonging to the mother and the fetus from the K normal distributions, and obtain the corresponding number N of free DNA fragments belonging to the mother and the fetus i , where i represents the number from 1 to K belonging to the mother and fetus;
[0037] S4: Perform three-dimensional linear fitting with the known and determined feta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com